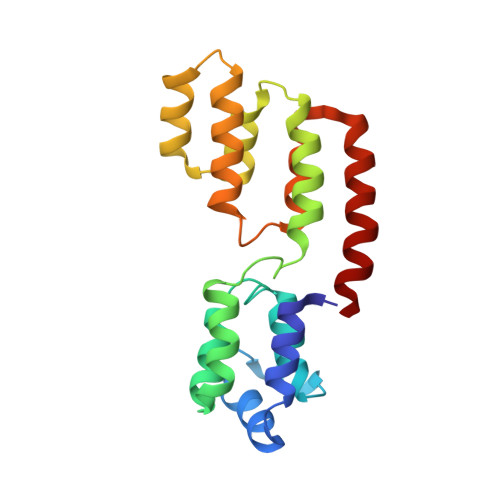
The Structure of FADD and Its Mode of Interaction with Procaspase-8
Carrington, P.E., Sandu, C., Wei, Y., Hill, J.M., Morisawa, G., Huang, T., Gavathiotis, E., Wei, Y., Werner, M.H.(2006) Mol Cell 22: 599-610
- PubMed: 16762833
- DOI: https://doi.org/10.1016/j.molcel.2006.04.018
- Primary Citation of Related Structures:
2GF5 - PubMed Abstract:
The structure of FADD has been solved in solution, revealing that the death effector domain (DED) and death domain (DD) are aligned with one another in an orthogonal, tail-to-tail fashion. Mutagenesis of FADD and functional reconstitution with its binding partners define the interaction with the intracellular domain of CD95 and the prodomain of procaspase-8 and reveal a self-association surface necessary to form a productive complex with an activated "death receptor." The identification of a procaspase-specific binding surface on the FADD DED suggests a preferential interaction with one, but not both, of the DEDs of procaspase-8 in a perpendicular arrangement. FADD self-association is mediated by a "hydrophobic patch" in the vicinity of F25 in the DED. The structure of FADD and its functional characterization, therefore, illustrate the architecture of key components in the death-inducing signaling complex.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, 1230 York Avenue, Box 42, New York, New York 10021, USA.