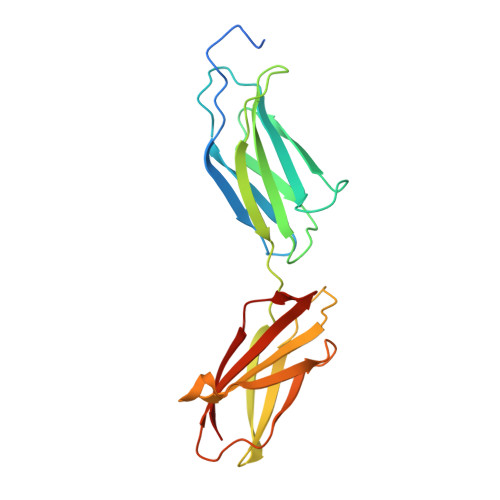
Structural insights into fibronectin type III domain-mediated signaling.
Bencharit, S., Cui, C.B., Siddiqui, A., Howard-Williams, E.L., Sondek, J., Zuobi-Hasona, K., Aukhil, I.(2007) J Mol Biol 367: 303-309
- PubMed: 17261313
- DOI: https://doi.org/10.1016/j.jmb.2006.10.017
- Primary Citation of Related Structures:
2GEE - PubMed Abstract:
The alternatively spliced type III extradomain B (EIIIB) of fibronectin (FN) is expressed only during embryogenesis, wound healing and tumorigenesis. The biological function of this domain is unclear. We describe here the first crystal structure of the interface between alternatively spliced EIIIB and its adjacent FN type III domain 8 (FN B-8). The opened CC' loop of EIIIB, and the rotation and tilt of EIIIB allow good access to the FG loop of FN-8, which is normally hindered by the CC' loop of FN-7. In addition, the AGEGIP sequence of the CC'' loop of EIIIB replaces the NGQQGN sequence of the CC' loop of FN-7. Finally, the CC'' loop of EIIIB forms an acidic groove with FN-8. These structural findings warrant future studies directed at identifying potential binding partners for FN B-8 interface, linking EIIIB to skeletal and cartilaginous development, wound healing, and tumorigenesis, respectively.
Organizational Affiliation:
Department of Prosthodontics, School of Dentistry, University of North Carolina, Chapel Hill, NC 27599, USA.