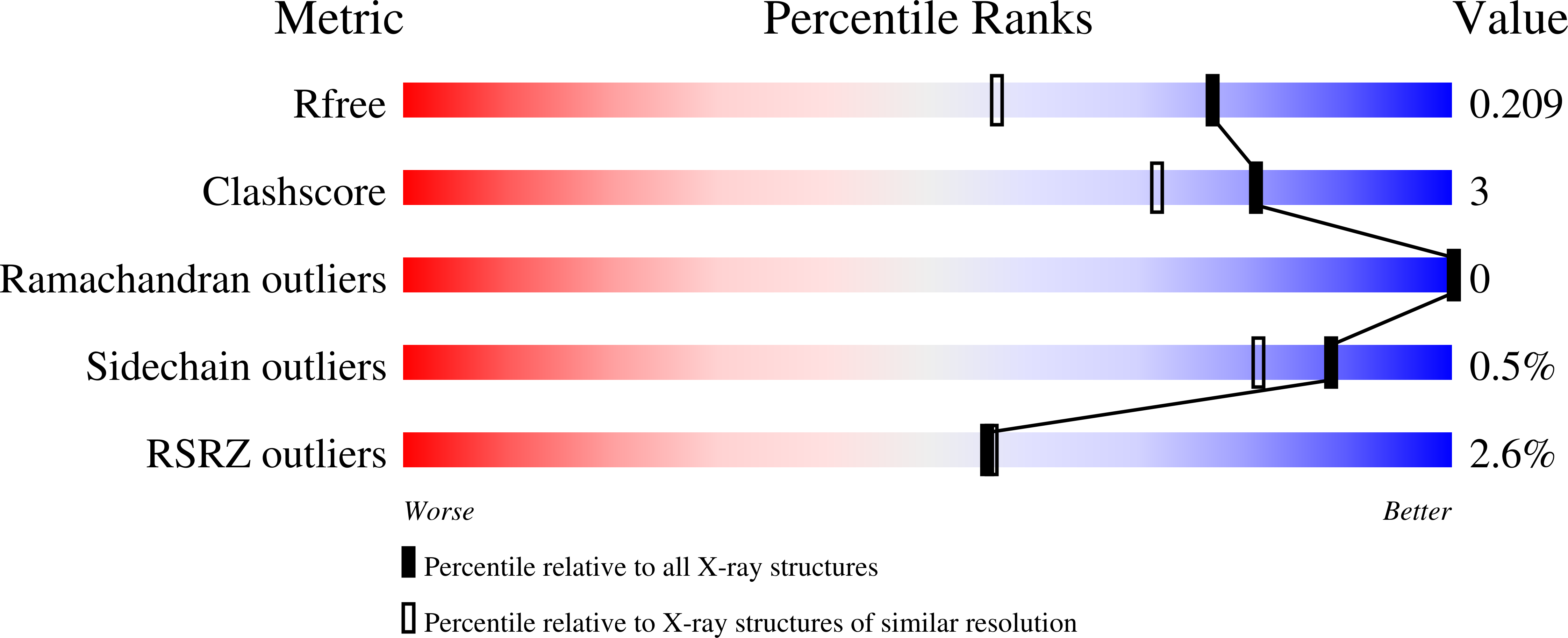
High-Affinity Inhibitors of Human NAD-Dependent 15-Hydroxyprostaglandin Dehydrogenase: Mechanisms of Inhibition and Structure-Activity Relationships.
Niesen, F.H., Schultz, L., Jadhav, A., Bhatia, C., Guo, K., Maloney, D.J., Pilka, E.S., Wang, M., Oppermann, U., Heightman, T.D., Simeonov, A.(2010) PLoS One 5: e13719-e13719
- PubMed: 21072165
- DOI: https://doi.org/10.1371/journal.pone.0013719
- Primary Citation of Related Structures:
2GDZ - PubMed Abstract:
15-Hydroxyprostaglandin dehydrogenase (15-PGDH, EC 1.1.1.141) is the key enzyme for the inactivation of prostaglandins, regulating processes such as inflammation or proliferation. The anabolic pathways of prostaglandins, especially with respect to regulation of the cyclooxygenase (COX) enzymes have been studied in detail; however, little is known about downstream events including functional interaction of prostaglandin-processing and -metabolizing enzymes. High-affinity probes for 15-PGDH will, therefore, represent important tools for further studies.
Organizational Affiliation:
Structural Genomics Consortium, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, United Kingdom.