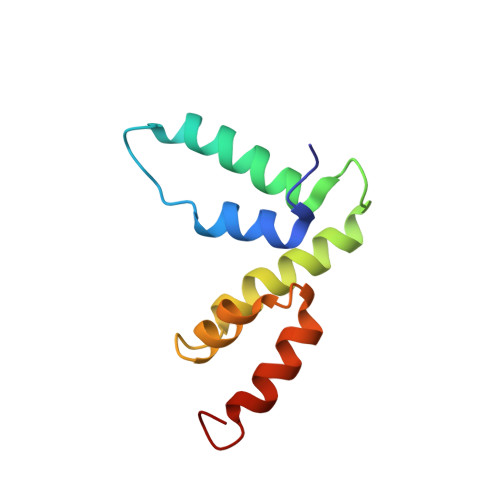
Structure of the SCAN domain from the tumor suppressor protein MZF1.
Peterson, F.C., Hayes, P.L., Waltner, J.K., Heisner, A.K., Jensen, D.R., Sander, T.L., Volkman, B.F.(2006) J Mol Biol 363: 137-147
- PubMed: 16950398
- DOI: https://doi.org/10.1016/j.jmb.2006.07.063
- Primary Citation of Related Structures:
2FI2 - PubMed Abstract:
The SCAN domain mediates interactions between members of a subfamily of zinc-finger transcription factors and is found in more than 60 C2H2 zinc finger genes in the human genome, including the tumor suppressor gene myeloid zinc finger 1 (MZF1). Glutathione-S-transferase pull-down assays showed that the MZF1 SCAN domain self-associates, and a Kd value of 600 nM was measured by intrinsic tryptophan fluorescence polarization. The MZF1 structure determined by NMR spectroscopy revealed a domain-swapped dimer. Each monomer consists of five alpha helices in two subdomains connected by the alpha2-alpha3 loop. Residues from helix 3 of each monomer compose the core of the dimer interface, while the alpha1-alpha2 loop and helix 2 pack against helices 3 and 5 from the opposing monomer. Comprehensive sequence analysis is coupled with the first high-resolution structure of a SCAN dimer to provide an initial view of the recognition elements that govern dimerization for this large family of transcription factors.
Organizational Affiliation:
Department of Biochemistry, Medical College of Wisconsin, 8701 Watertown Plank Road Milwaukee, WI 53226, USA.