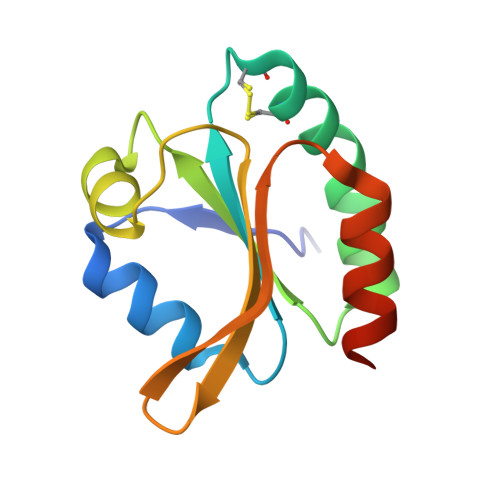
High-resolution structure of recombinant Trichomonas vaginalis thioredoxin.
Iulek, J., Alphey, M.S., Westrop, G.D., Coombs, G.H., Hunter, W.N.(2006) Acta Crystallogr D Biol Crystallogr 62: 216-220
- PubMed: 16421453
- DOI: https://doi.org/10.1107/S0907444905039946
- Primary Citation of Related Structures:
2F51 - PubMed Abstract:
The structure of thioredoxin from the anaerobic organism Trichomonas vaginalis (TvTrx) has been determined at 1.9 angstroms resolution. The structure is that of a typical thioredoxin: a five-stranded beta-sheet structure with two alpha-helices on either side. The active site of the protein carries a Trp-Cys-Gly-Pro-Cys motif, residues 34-38, at the N-terminus of an alpha-helix (alpha2). The cysteine residues in this motif form a redox-active disulfide necessary for thioredoxin activity. With high-resolution data available, it was possible to model numerous amino-acid side chains in alternate conformations and this includes the redox-active disulfide cysteine residues. The sample was initially in the oxidized state and the use of X-rays from an intense third-generation synchrotron source resulted in partial photoreduction of this labile redox centre. Comparisons with previously determined thioredoxin structures indicate that TvTrx is most similar to the human homologue, although the insertion of three residues between strands beta4 and beta5 makes the corresponding turn longer and more flexible in TvTrx. In addition, three significant amino-acid differences are identified on the protein surfaces near to the active-site Cys35. These residues may contribute to the interactions that specific thioredoxins form with their cognate physiological partners.
Organizational Affiliation:
Department of Chemistry, Biotechnology Center, State University of Ponta Grossa, Ponta Grossa, Paraná, Brazil.