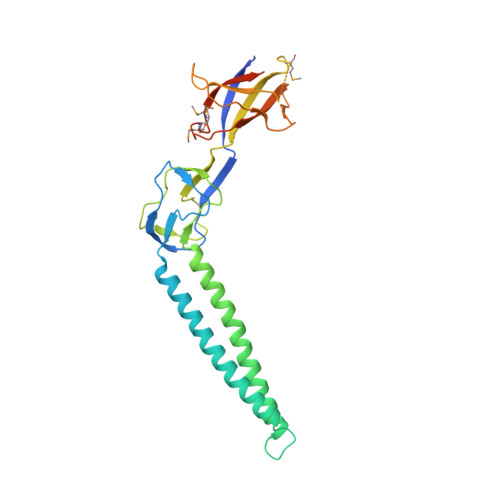
Conformational Flexibility in the Multidrug Efflux System Protein AcrA.
Mikolosko, J., Bobyk, K., Zgurskaya, H.I., Ghosh, P.(2006) Structure 14: 577-587
- PubMed: 16531241
- DOI: https://doi.org/10.1016/j.str.2005.11.015
- Primary Citation of Related Structures:
2F1M - PubMed Abstract:
Intrinsic resistance to multiple drugs in many gram-negative bacterial pathogens is conferred by resistance nodulation cell division efflux pumps, which are composed of three essential components as typified by the extensively characterized Escherichia coli AcrA-AcrB-TolC system. The inner membrane drug:proton antiporter AcrB and the outer membrane channel TolC export chemically diverse compounds out of the bacterial cell, and require the activity of the third component, the periplasmic protein AcrA. The crystal structures of AcrB and TolC have previously been determined, and we complete the molecular picture of the efflux system by presenting the structure of a stable fragment of AcrA. The AcrA fragment resembles the elongated sickle shape of its homolog Pseudomonas aeruginosa MexA, being composed of three domains: beta-barrel, lipoyl, and alpha-helical hairpin. Notably, unsuspected conformational flexibility in the alpha-helical hairpin domain of AcrA is observed, which has potential mechanistic significance in coupling between AcrA conformations and TolC channel opening.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92037, USA.