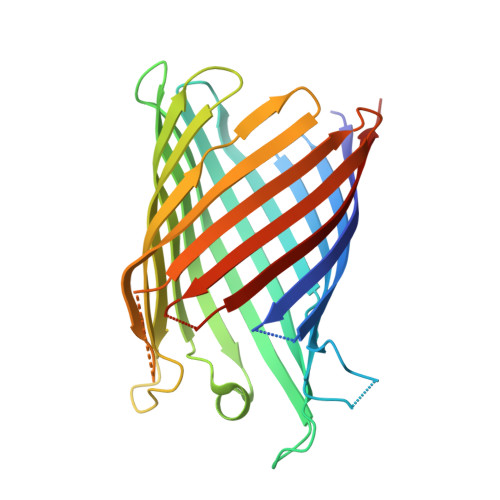
Crystal Structure of the Monomeric Porin OmpG.
Subbarao, G.V., van den Berg, B.(2006) J Mol Biol 360: 750-759
- PubMed: 16797588
- DOI: https://doi.org/10.1016/j.jmb.2006.05.045
- Primary Citation of Related Structures:
2F1C - PubMed Abstract:
The outer membrane (OM) of Gram-negative bacteria contains a large number of channel proteins that mediate the uptake of ions and nutrients necessary for growth and functioning of the cell. An important group of OM channel proteins are the porins, which mediate the non-specific, diffusion-based passage of small (<600 Da) polar molecules. All porins of Gram-negative bacteria that have been crystallized to date form stable trimers, with each monomer composed of a 16-stranded beta-barrel with a relatively narrow central pore. In contrast, the OmpG porin is unique, as it appears to function as a monomer. We have determined the X-ray crystal structure of OmpG from Escherichia coli to a resolution of 2.3 A. The structure shows a 14-stranded beta-barrel with a relatively simple architecture. Due to the absence of loops that fold back into the channel, OmpG has a large ( approximately 13 A) central pore that is considerably wider than those of other E. coli porins, and very similar in size to that of the toxin alpha-hemolysin. The architecture of the channel, together with previous biochemical and other data, suggests that OmpG may form a non-specific channel for the transport of larger oligosaccharides. The structure of OmpG provides the starting point for engineering studies aiming to generate selective channels and for the development of biosensors.
Organizational Affiliation:
Program in Molecular Medicine, University of Massachusetts Medical School, 373 Plantation Street, Worcester, MA 01605, USA.