The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition
Kamtekar, S., Berman, A.J., Wang, J., Lazaro, J.M., de Vega, M., Blanco, L., Salas, M., Steitz, T.A.(2006) EMBO J 25: 1335-1343
- PubMed: 16511564
- DOI: https://doi.org/10.1038/sj.emboj.7601027
- Primary Citation of Related Structures:
2EX3 - PubMed Abstract:
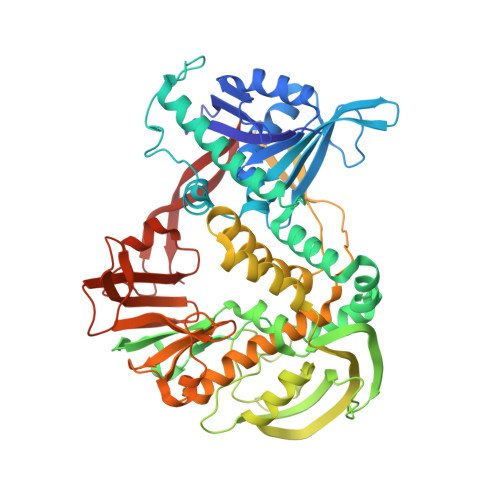
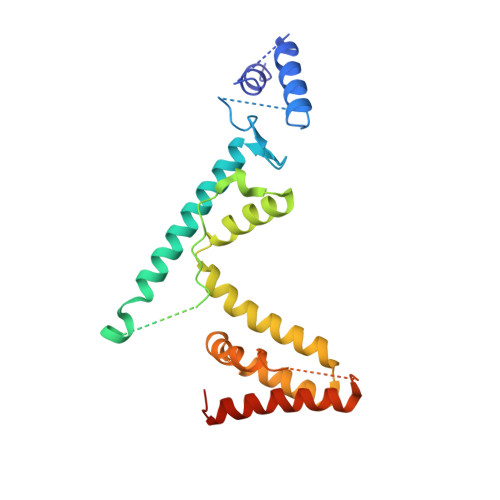
The absolute requirement for primers in the initiation of DNA synthesis poses a problem for replicating the ends of linear chromosomes. The DNA polymerase of bacteriophage phi29 solves this problem by using a serine hydroxyl of terminal protein to prime replication. The 3.0 A resolution structure shows one domain of terminal protein making no interactions, a second binding the polymerase and a third domain containing the priming serine occupying the same binding cleft in the polymerase as duplex DNA does during elongation. Thus, the progressively elongating DNA duplex product must displace this priming domain. Further, this heterodimer of polymerase and terminal protein cannot accommodate upstream template DNA, thereby explaining its specificity for initiating DNA synthesis only at the ends of the bacteriophage genome. We propose a model for the transition from the initiation to the elongation phases in which the priming domain of terminal protein moves out of the active site as polymerase elongates the primer strand. The model indicates that terminal protein should dissociate from polymerase after the incorporation of approximately six nucleotides.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8114, USA.