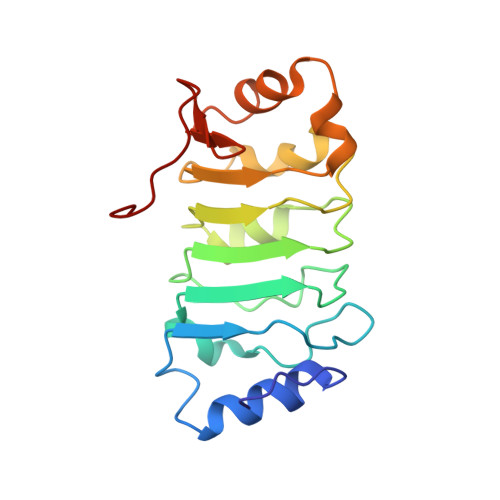
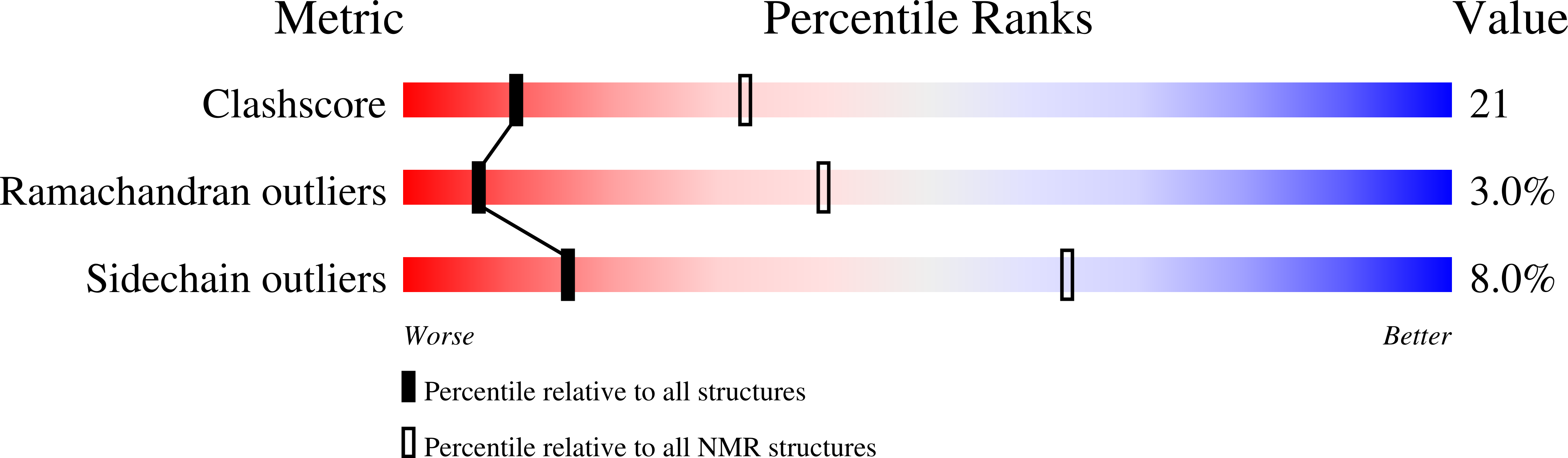Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
Tochio, N., Umehara, T., Munemasa, Y., Suzuki, T., Sato, S., Tsuda, K., Koshiba, S., Kigawa, T., Nagai, R., Yokoyama, S.(2010) J Mol Biol 401: 97-114
- PubMed: 20538007
- DOI: https://doi.org/10.1016/j.jmb.2010.06.005
- Primary Citation of Related Structures:
2ELL, 2RR6 - PubMed Abstract:
Eukaryotic gene expression is regulated by histone deposition onto and eviction from nucleosomes, which are mediated by several chromatin-modulating factors. Among them, histone chaperones are key factors that facilitate nucleosome assembly. Acidic nuclear phosphoprotein 32B (ANP32B) belongs to the ANP32 family, which shares N-terminal leucine-rich repeats (LRRs) and a C-terminal variable anionic region. The C-terminal region functions as an inhibitor of histone acetylation, but the functional roles of the LRR domain in chromatin regulation have remained elusive. Here, we report that the LRR domain of ANP32B possesses histone chaperone activity and forms a curved structure with a parallel beta-sheet on the concave side and mostly helical elements on the convex side. Our analyses revealed that the interaction of ANP32B with the core histones H3-H4 occurs on its concave side, and both the acidic and hydrophobic residues that compose the concave surface are critical for histone binding. These results provide a structural framework for understanding the functional mechanisms of acidic histone chaperones.
Organizational Affiliation:
RIKEN Systems and Structural Biology Center, 1-7-22 Suehiro-cho, Tsurumi, Yokohama 230-0045, Japan.