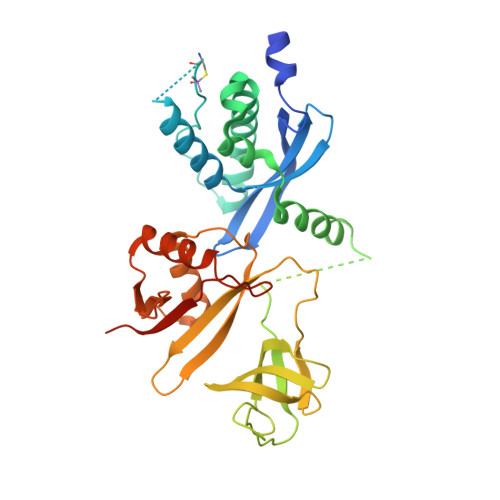
Full-length p40phox structure suggests a basis for regulation mechanism of its membrane binding.
Honbou, K., Minakami, R., Yuzawa, S., Takeya, R., Suzuki, N.N., Kamakura, S., Sumimoto, H., Inagaki, F.(2007) EMBO J 26: 1176-1186
- PubMed: 17290225
- DOI: https://doi.org/10.1038/sj.emboj.7601561
- Primary Citation of Related Structures:
2DYB - PubMed Abstract:
The superoxide-producing phagocyte NADPH oxidase is activated during phagocytosis to destroy ingested microbes. The adaptor protein p40phox associates via the PB1 domain with the essential oxidase activator p67phox, and is considered to function by recruiting p67phox to phagosomes; in this process, the PX domain of p40phox binds to phosphatidylinositol 3-phosphate [PtdIns(3)P], a lipid abundant in the phagosomal membrane. Here we show that the PtdIns(3)P-binding activity of p40phox is normally inhibited by the PB1 domain both in vivo and in vitro. The crystal structure of the full-length p40phox reveals that the inhibition is mediated via intramolecular interaction between the PB1 and PX domains. The interface of the p40phox PB1 domain for the PX domain localizes on the opposite side of that for the p67phox PB1 domain, and thus the PB1-mediated PX regulation occurs without preventing the PB1-PB1 association with p67phox.
Organizational Affiliation:
Laboratory of Structural Biology, Graduate School of Pharmaceutical Sciences, Hokkaido University, Sapporo, Japan.