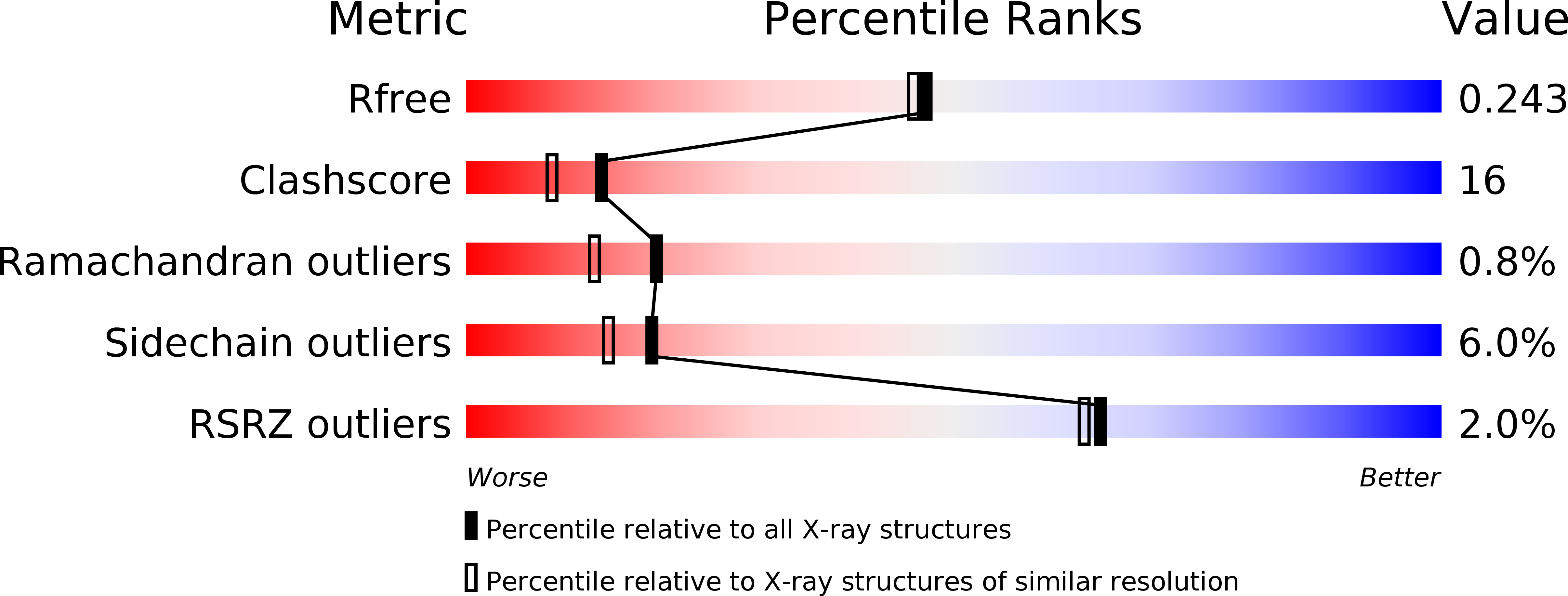
Structure of the minimized alpha/beta-hydrolase fold protein from Thermus thermophilus HB8.
Xie, Y., Takemoto, C., Kishishita, S., Uchikubo-Kamo, T., Murayama, K., Chen, L., Liu, Z.J., Wang, B.C., Manzoku, M., Ebihara, A., Kuramitsu, S., Shirouzu, M., Yokoyama, S.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 993-997
- PubMed: 18084077
- DOI: https://doi.org/10.1107/S1744309107061106
- Primary Citation of Related Structures:
2DST - PubMed Abstract:
The gene encoding TTHA1544 is a singleton found in the Thermus thermophilus HB8 genome and encodes a 131-amino-acid protein. The crystal structure of TTHA1544 has been determined at 2.0 A resolution by the single-wavelength anomalous dispersion method in order to elucidate its function. There are two molecules in the asymmetric unit. Each molecule consists of four alpha-helices and six beta-strands, with the beta-strands composing a central beta-sheet. A structural homology search revealed that the overall structure of TTHA1544 resembles the alpha/beta-hydrolase fold, although TTHA1544 lacks the catalytic residues of a hydrolase. These results suggest that TTHA1544 represents the minimized alpha/beta-hydrolase fold and that an additional component would be required for its activity.
Organizational Affiliation:
Protein Research Group, Genomic Sciences Center, RIKEN Yokohama Institute, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa 230-0045, Japan.