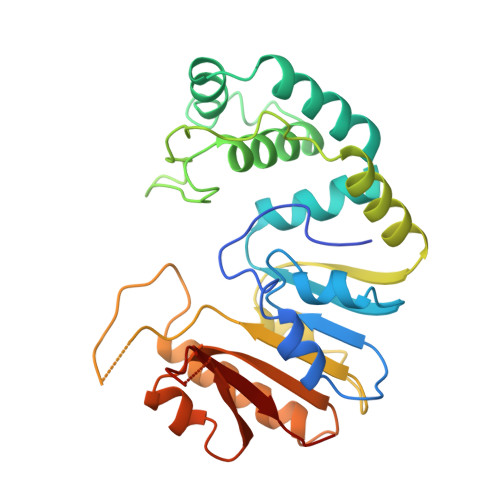
Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine.
Tran, P.H., Korszun, Z.R., Cerritelli, S., Springhorn, S.S., Lacks, S.A.(1998) Structure 6: 1563-1575
- PubMed: 9862809
- DOI: https://doi.org/10.1016/s0969-2126(98)00154-3
- Primary Citation of Related Structures:
2DPM - PubMed Abstract:
. Methyltransferases (Mtases) catalyze the transfer of methyl groups from S-adenosylmethionine (AdoMet) to a variety of small molecular and macromolecular substrates. These enzymes contain a characteristic alpha/beta structural fold. Four groups of DNA Mtases have been defined and representative structures have been determined for three groups. DpnM is a DNA Mtase that acts on adenine N6 in the sequence GATC; the enzyme represents group alpha DNA Mtases, for which no structures are known.
Organizational Affiliation:
Department of Biology, Brookhaven National Laboratory, Upton, NY 11973,USA.