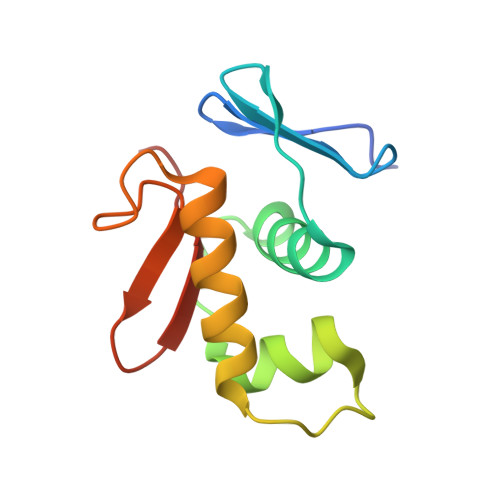
Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Okajima, T., Doi, A., Okada, A., Gotoh, Y., Tanizawa, K., Utsumi, R.(2008) FEBS Lett 582: 3434-3438
- PubMed: 18789936
- DOI: https://doi.org/10.1016/j.febslet.2008.09.007
- Primary Citation of Related Structures:
2D1V - PubMed Abstract:
A response regulator YycF and its cognate sensor kinase YycG constitute the two-component signal transduction system essential for growth of Gram-positive bacteria with a low GC content. We have determined the X-ray crystal structure of the effector domain of Bacillus subtilis YycF involved in DNA binding. The structure, containing a winged helix-turn-helix motif, was found to be very similar to that of the response regulator PhoB from Escherichia coli. Specific binding of YycF to the PhoB-regulated alkaline phosphatase promoter was also demonstrated.
Organizational Affiliation:
Institute of Scientific and Industrial Research, Osaka University, Ibaraki, Osaka 567-0047, Japan. tokajima@sanken.osaka-u.ac.jp