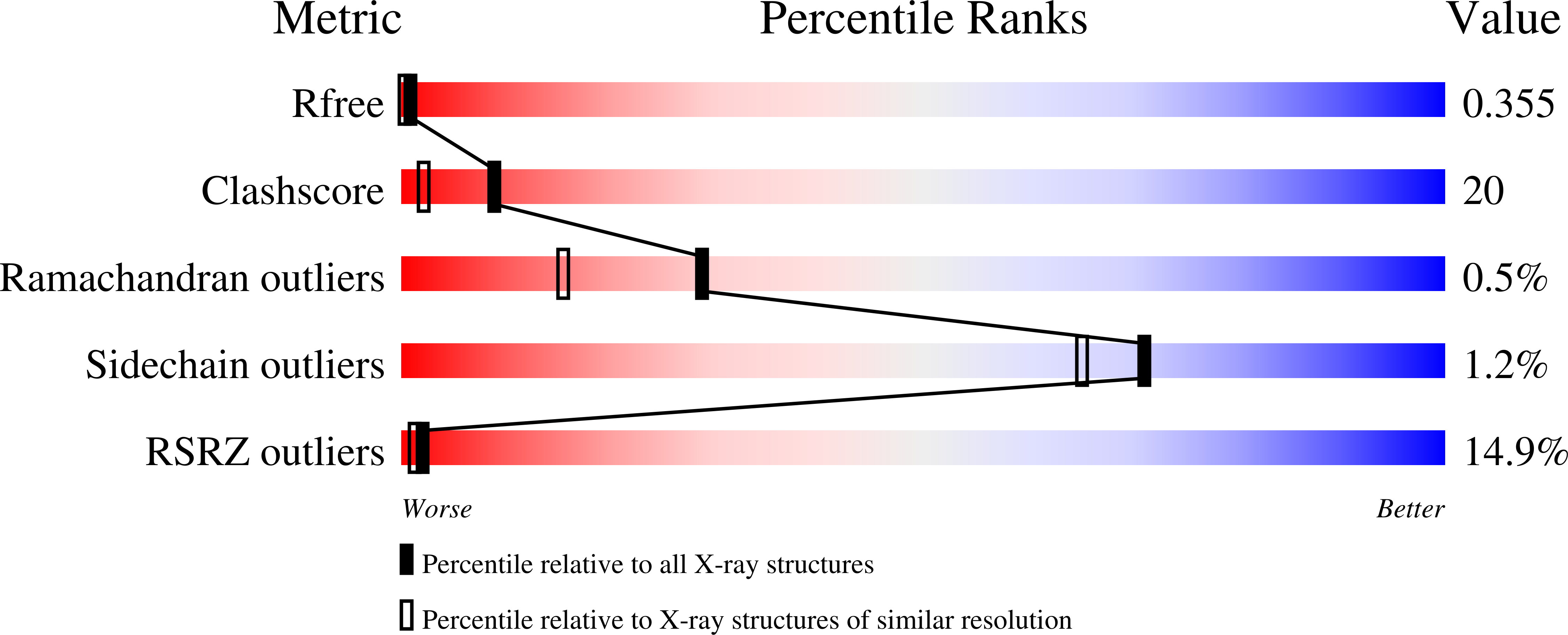
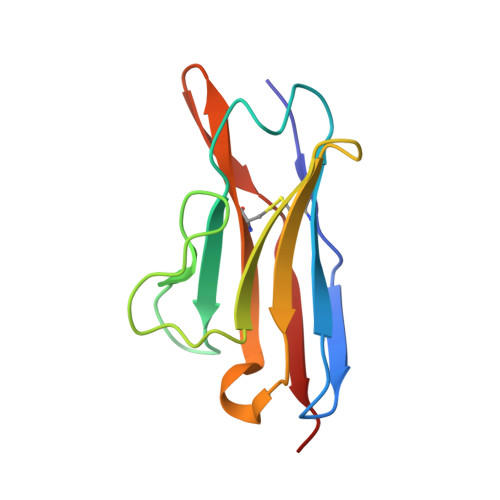
Tertiary structure of human lambda 6 light chains.
Pokkuluri, P.R., Solomon, A., Weiss, D.T., Stevens, F.J., Schiffer, M.(1999) Amyloid 6: 165-171
- PubMed: 10524280
- DOI: https://doi.org/10.3109/13506129909007322
- Primary Citation of Related Structures:
1CD0, 2CD0 - PubMed Abstract:
AL amyloidosis is a disease process characterized by the pathologic deposition of monoclonal light chains in tissue. To date, only limited information has been obtained on the molecular features that render such light chains amyloidogenic. Although protein products of the major human V kappa and V lambda gene families have been identified in AL deposits, one particular subgroup--lambda 6--has been found to be preferentially associated with this disease. Notably, the variable region of lambda 6 proteins (V lambda 6) has distinctive primary structural features including the presence in the third framework region (FR3) of two additional amino acid residues that distinguish members of this subgroup from other types of light chains. However, the structural consequences of these alterations have not been elucidated. To determine if lambda 6 proteins possess unique tertiary structural features, as compared to light chains of other V lambda subgroups, we have obtained x-ray diffraction data on crystals prepared from two recombinant V lambda 6 molecules. These components, isolated from a bacterial expression system, were generated from lambda 6-related cDNAs cloned from bone marrow-derived plasma cells from a patient (Wil) who had documented AL amyloidosis and another (Jto) with multiple myeloma and tubular cast nephropathy, but no evident fibrillar deposits. The x-ray crystallographic analyses revealed that the two-residue insertion located between positions 68 and 69 (not between 66 and 67 as previously surmised) extended an existing loop region that effectively increased the surface area adjacent to the first complementarity determining region (CDR1). Further, an unusual interaction between the Arg 25 and Phe 2 residues commonly found in lambda 6 molecules was noted. However, the structures of V lambda 6 Wil and Jto also differed from each other, as evidenced by the presence in the latter of certain ionic and hydrophobic interactions that we posit increased protein stability and thus prevented amyloid formation.