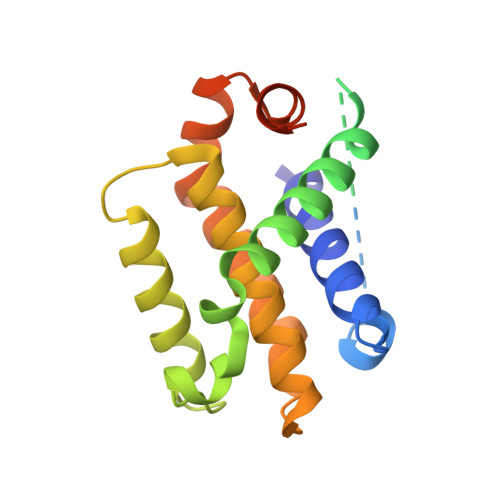
Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral Bcl-2 of Murine Gamma-Herpesvirus 68.
Ku, B., Woo, J., Liang, C., Lee, K., Hong, H., Xiaofeni, E., Kim, K., Jung, J.U., Oh, B.(2008) PLoS Pathog 4: E25
- PubMed: 18248095
- DOI: https://doi.org/10.1371/journal.ppat.0040025
- Primary Citation of Related Structures:
2BZW, 3BL2 - PubMed Abstract:
All gammaherpesviruses express homologues of antiapoptotic B-cell lymphoma-2 (BCL-2) to counter the clearance of infected cells by host antiviral defense machineries. To gain insights into the action mechanisms of these viral BCL-2 proteins, we carried out structural and biochemical analyses on the interactions of M11, a viral BCL-2 of murine gamma-herpesvirus 68, with a fragment of proautophagic Beclin1 and BCL-2 homology 3 (BH3) domain-containing peptides derived from an array of proapoptotic BCL-2 family proteins. Mainly through hydrophobic interactions, M11 bound the BH3-like domain of Beclin1 with a dissociation constant of 40 nanomole, a markedly tighter affinity compared to the 1.7 micromolar binding affinity between cellular BCL-2 and Beclin1. Consistently, M11 inhibited autophagy more efficiently than BCL-2 in NIH3T3 cells. M11 also interacted tightly with a BH3 domain peptide of BAK and those of the upstream BH3-only proteins BIM, BID, BMF, PUMA, and Noxa, but weakly with that of BAX. These results collectively suggest that M11 potently inhibits Beclin1 in addition to broadly neutralizing the proapoptotic BCL-2 family in a similar but distinctive way from cellular BCL-2, and that the Beclin1-mediated autophagy may be a main target of the virus.
Organizational Affiliation:
Division of Molecular and Life Sciences, Center for Biomolecular Recognition, Pohang University of Science and Technology, Pohang, Kyungbuk, Korea.