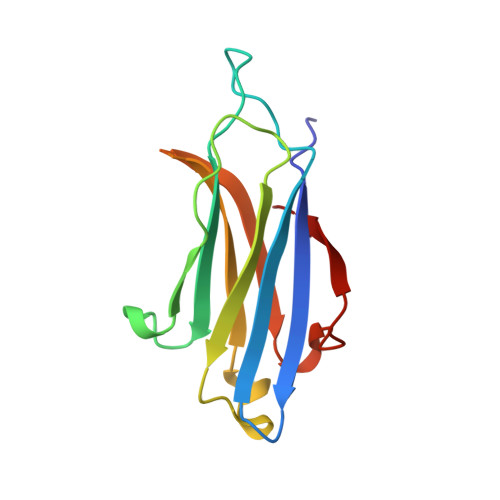
Crystal Structure of the Rim2 C(2)A-Domain at 1.4 A Resolution.
Dai, H., Tomchick, D.R., Garcia, J., Sudhof, T.C., Machius, M., Rizo, J.(2005) Biochemistry 44: 13533
- PubMed: 16216076
- DOI: https://doi.org/10.1021/bi0513608
- Primary Citation of Related Structures:
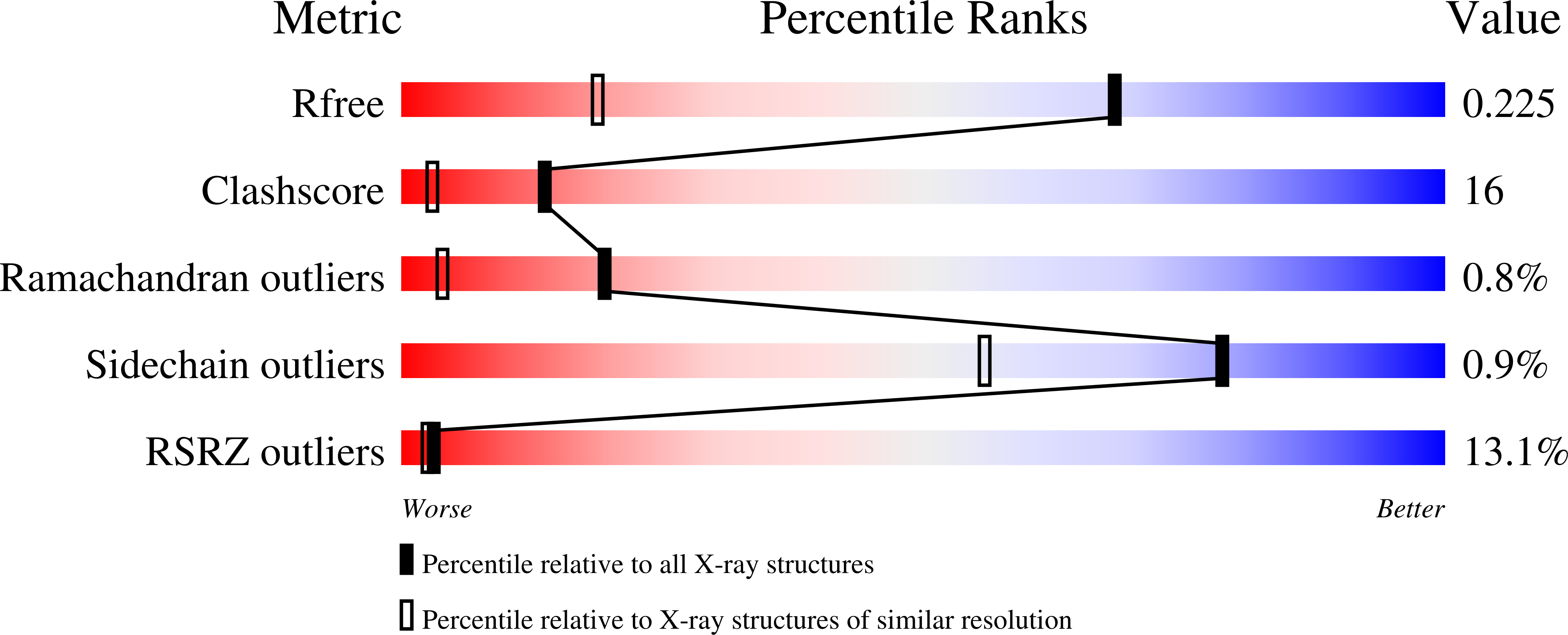
2BWQ - PubMed Abstract:
RIMs are large proteins that contain two C2-domains and are localized at presynaptic active zones, where neurotransmitters are released. RIMs play key roles in synaptic vesicle priming and regulation of presynaptic plasticity. A mutation in the RIM1 C2A-domain has been implicated in autosomal dominant cone-rod dystrophy (CORD7). The RIM C2A-domain does not contain the full complement of aspartate residues that commonly mediate Ca2+ binding at the top loops of C2-domains, and has been reported to interact with SNAP-25 and synaptotagmin 1, two proteins from the Ca2+-dependent membrane fusion machinery. Here we have used NMR spectroscopy and X-ray crystallography to analyze the structure and biochemical properties of the RIM2 C2A-domain, which is closely related to the RIM1 C2A-domain. We find that the RIM2 C2A-domain does not bind Ca2+. Moreover, little binding of the RIM2 C2A-domain to SNAP-25 and to the C2-domains of synaptotagmin 1 was detected by NMR experiments, suggesting that as yet unidentified interactions of the RIM C2A-domain mediate its function. The crystal structure of the RIM2 C2A-domain using data to 1.4 A resolution reveals a beta-sandwich that resembles those observed for other C2-domains, but exhibits a unique dipolar distribution of electrostatic charges whereby one edge of the beta-sandwich is highly positive and the other edge is highly negative. The location of the mutation site implicated in CORD7 at the bottom of the domain and the pattern of sequence conservation suggest that, in contrast to most C2-domains, the RIM C2A-domains may function through Ca2+-independent interactions involving their bottom face.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, Texas 75390, USA.