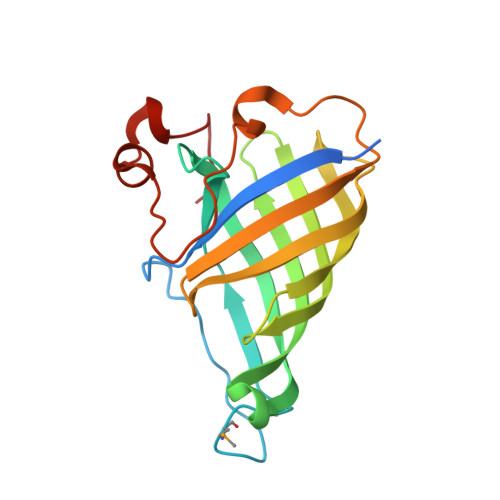
The Crystal Structure of Arabidopsis Thaliana Allene Oxide Cyclase: Insights Into the Oxylipin Cyclization Reaction.
Hofmann, E., Zerbe, P., Schaller, F.(2006) Plant Cell 18: 3201
- PubMed: 17085685
- DOI: https://doi.org/10.1105/tpc.106.043984
- Primary Citation of Related Structures:
2BRJ, 2DIO, 2GIN - PubMed Abstract:
We describe the crystallization and structure elucidation of Arabidopsis thaliana allene oxide cyclase 2 (AOC2), a key enzyme in the biosynthesis of jasmonates. In a coupled reaction with allene oxide synthase, AOC2 releases the first cyclic and biologically active metabolite, 12-oxo-phytodienoic acid (OPDA). AOC2 (AT3G25770) folds into an eight-stranded antiparallel beta-barrel with a C-terminal partial helical extension. The protein forms a hydrophobic binding cavity with two distinct polar patches. AOC2 is trimeric in crystals, in vitro and in planta. Based on the observed folding pattern, we assigned AOC2 as a low molecular weight member of the lipocalin family with enzymatic activity in plants. We determined the binding position of the competitive inhibitor vernolic acid (a substrate analog) in the binding pocket. Based on models for bound substrate 12,13-epoxy-9,11,15-octadecatrienoic acid and product OPDA, we propose a reaction scheme that explains the influence of the C15 double bond on reactivity. Reaction is promoted by anchimeric assistance through a conserved Glu residue. The transition state with a pentadienyl carbocation and an oxyanion is stabilized by a strongly bound water molecule and favorable pi-pi interactions with aromatic residues in the cavity. Stereoselectivity results from steric restrictions to the necessary substrate isomerizations imposed by the protein.
Organizational Affiliation:
Lehrstuhl für Biophysik, Ruhr-Universität Bochum, D-44780 Bochum, Germany. eckhard.hofmann@bph.rub.de