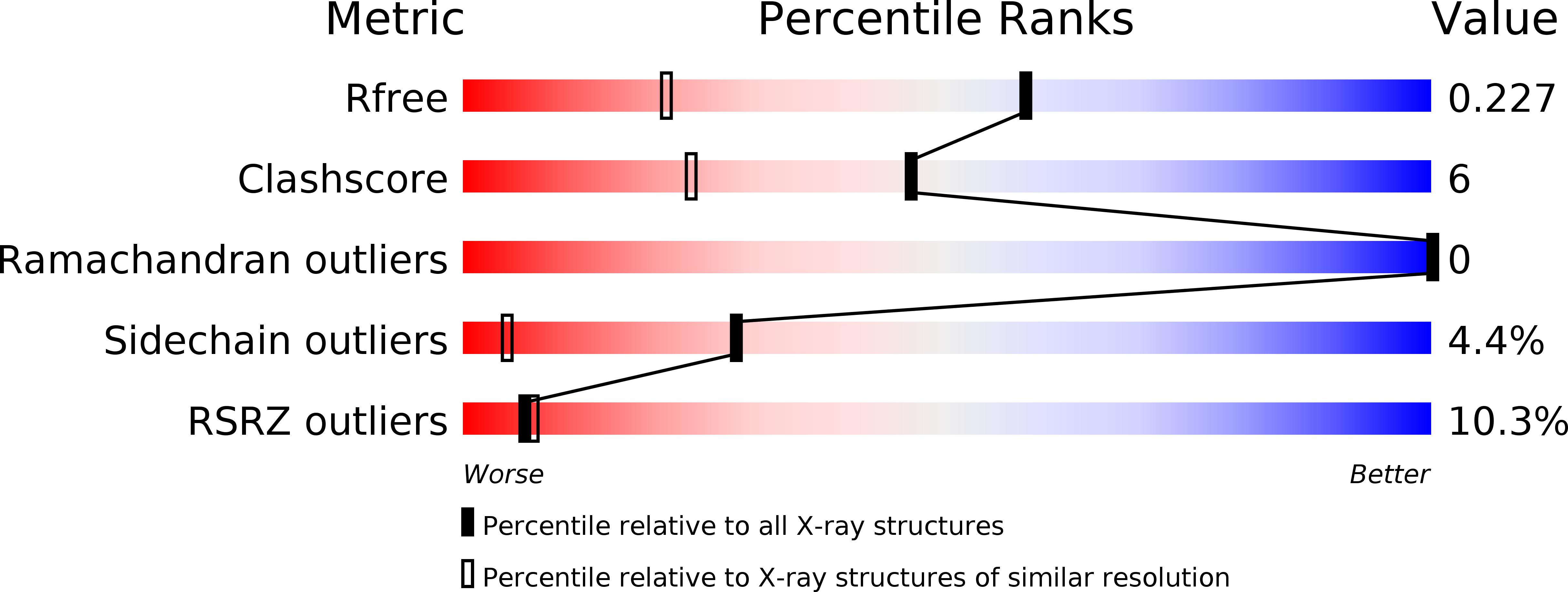
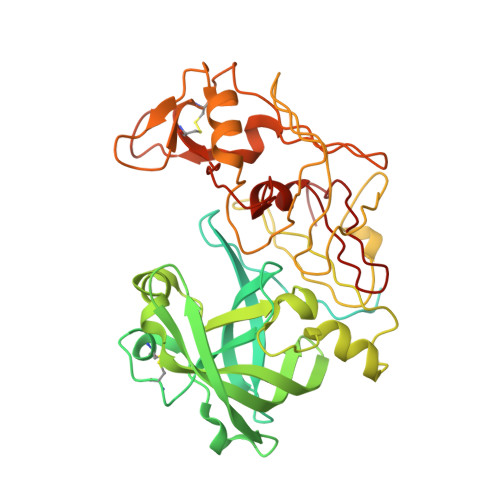
X-Ray Structure of Plasmepsin II Complexed with a Potent Achiral Inhibitor.
Prade, L., Jones, A.F., Boss, C., Richard-Bildstein, S., Meyer, S., Binkert, C., Bur, D.(2005) J Biol Chem 280: 23837
- PubMed: 15840589
- DOI: https://doi.org/10.1074/jbc.M501519200
- Primary Citation of Related Structures:
2BJU - PubMed Abstract:
The malaria parasite Plasmodium falciparum degrades host cell hemoglobin inside an acidic food vacuole during the blood stage of the infectious cycle. A number of aspartic proteinases called plasmepsins (PMs) have been identified to play important roles in this degradation process and therefore generated significant interest as new antimalarial targets. Several x-ray structures of PMII have been described previously, but thus far, structure-guided drug design has been hampered by the fact that only inhibitors comprising a statine moiety or derivatives thereof have been published. Our drug discovery efforts to find innovative, cheap, and easily synthesized inhibitors against aspartic proteinases yielded some highly potent non-peptidic achiral inhibitors. A highly resolved (1.6 A) x-ray structure of PMII is presented, featuring a potent achiral inhibitor in an unprecedented orientation, contacting the catalytic aspartates indirectly via the "catalytic" water. Major side chain rearrangements in the active site occur, which open up a new pocket and allow a new binding mode of the inhibitor. Moreover, a second inhibitor molecule could be located unambiguously in the active site of PMII. These newly obtained structural insights will further guide our attempts to improve compound properties eventually leading to the identification of molecules suitable as antimalarial drugs.
Organizational Affiliation:
Actelion Pharmaceuticals Ltd., Gewerbestrasse 16, CH-4123 Allschwil, Switzerland.