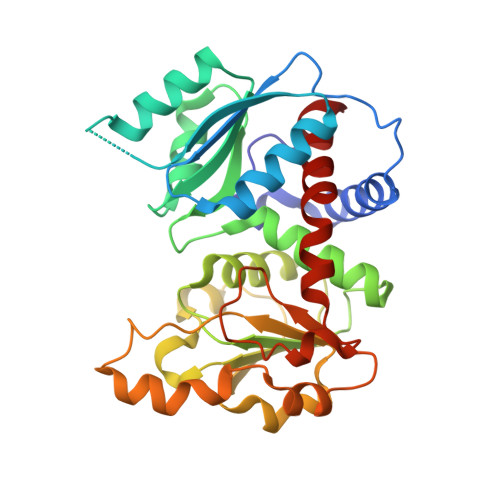
Crystal structure of Sulfolobus acidocaldarius aspartate carbamoyltransferase in complex with its allosteric activator CTP.
De Vos, D., Xu, Y., Aerts, T., Van Petegem, F., Van Beeumen, J.J.(2008) Biochem Biophys Res Commun 372: 40-44
- PubMed: 18477471
- DOI: https://doi.org/10.1016/j.bbrc.2008.04.173
- Primary Citation of Related Structures:
2BE9 - PubMed Abstract:
Aspartate carbamoyltransferase (ATCase) is a paradigm for allosteric regulation of enzyme activity. B-class ATCases display very similar homotropic allosteric behaviour, but differ extensively in their heterotropic patterns. The ATCase from the thermoacidophilic archaeon Sulfolobus acidocaldarius, for example, is strongly activated by its metabolic pathway's end product CTP, in contrast with Escherichia coli ATCase which is inhibited by CTP. To investigate the structural basis of this property, we have solved the crystal structure of the S. acidocaldarius enzyme in complex with CTP. Structure comparison reveals that effector binding does not induce similar large-scale conformational changes as observed for the E. coli ATCase. However, shifts in sedimentation coefficients upon binding of the bi-substrate analogue PALA show the existence of structurally distinct allosteric states. This suggests that the so-called "Nucleotide-Perturbation model" for explaining heterotropic allosteric behaviour, which is based on global conformational strain, is not a general mechanism of B-class ATCases.
Organizational Affiliation:
Laboratory of Protein Biochemistry and Protein Engineering, Ghent University, K.L. Ledeganckstraat 35, 9000 Gent, Belgium.