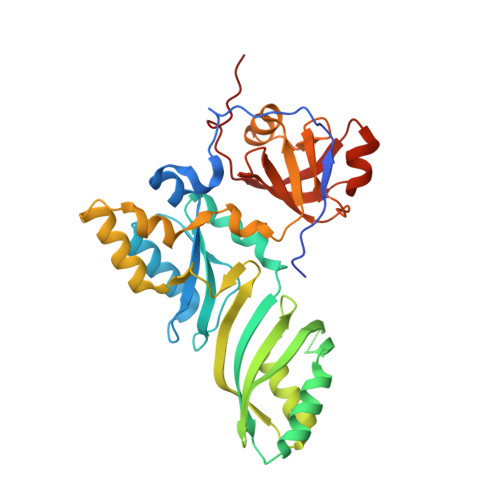
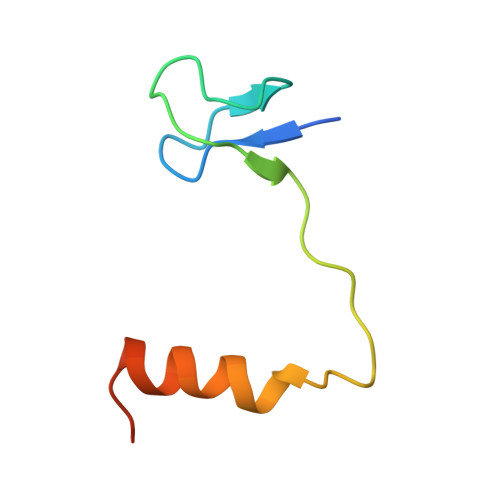
Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity
Manival, X., Charron, C., Fourmann, J.B., Godard, F., Charpentier, B., Branlant, C.(2006) Nucleic Acids Res 34: 826-839
- PubMed: 16456033
- DOI: https://doi.org/10.1093/nar/gkj482
- Primary Citation of Related Structures:
2AUS - PubMed Abstract:
In archaeal rRNAs, the isomerization of uridine into pseudouridine (Psi) is achieved by the H/ACA sRNPs and the minimal set of proteins required for RNA:Psi-synthase activity is the aCBF5-aNOP10 protein pair. The crystal structure of the aCBF5-aNOP10 heterodimer from Pyrococcus abyssi was solved at 2.1 A resolution. In this structure, protein aNOP10 has an extended shape, with a zinc-binding motif at the N-terminus and an alpha-helix at the C-terminus. Both motifs contact the aCBF5 catalytic domain. Although less efficiently as does the full-length aNOP10, the aNOP10 C-terminal domain binds aCBF5 and stimulates the RNA-guided activity. We show that the C-terminal domain of aCBF5 (the PUA domain), which is wrapped by an N-terminal extension of aCBF5, plays a crucial role for aCBF5 binding to the guide sRNA. Addition of this domain in trans partially complement particles assembled with an aCBF5DeltaPUA truncated protein. In the crystal structure, the aCBF5-aNOP10 complex forms two kinds of heterotetramers with parallel and perpendicular orientations of the aNOP10 terminal alpha-helices, respectively. By gel filtration assay, we showed that aNOP10 can dimerize in solution. As both residues Y41 and L48 were needed for dimerization, the dimerization likely takes place by interaction of parallel alpha-helices.
Organizational Affiliation:
Laboratoire de Maturation des ARN et Enzymologie Moléculaire, UMR 7567 UHP-CNRS, Université des Sciences et Techniques Henri Poincaré, Nancy I, 54506 Vandoeuvre-Lès-Nancy cedex, France.