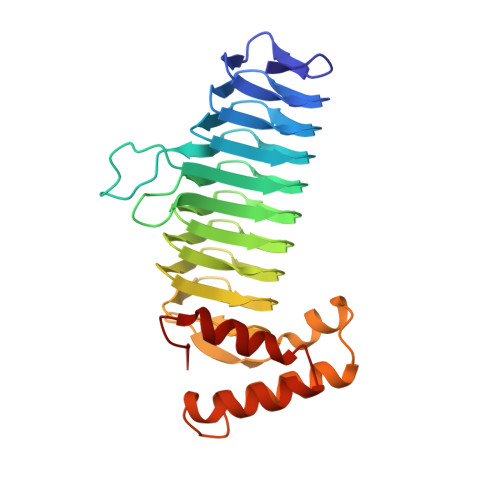
Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide.
Williams, A.H., Immormino, R.M., Gewirth, D.T., Raetz, C.R.(2006) Proc Natl Acad Sci U S A 103: 10877-10882
- PubMed: 16835299
- DOI: https://doi.org/10.1073/pnas.0604465103
- Primary Citation of Related Structures:
2AQ9 - PubMed Abstract:
UDP-GlcNAc acyltransferase (LpxA) catalyzes the first step of lipid A biosynthesis, the transfer of the R-3-hydroxyacyl chain from R-3-hydroxyacyl acyl carrier protein (ACP) to the glucosamine 3-OH group of UDP-GlcNAc. LpxA is essential for the growth of Escherichia coli and related Gram-negative bacteria. The crystal structure of the E. coli LpxA homotrimer, determined previously at 2.6 A in the absence of substrates or inhibitors, revealed that LpxA contains an unusual, left-handed parallel beta-helix fold. We now present the crystal structure at 1.8 A resolution of E. coli LpxA in a complex with a pentadecapeptide, peptide 920. Three peptides, each of which adopts a beta-hairpin conformation, are bound per LpxA trimer. The peptides are located at the interfaces of adjacent subunits in the vicinity of the three active sites. Each peptide interacts with residues from both adjacent subunits. Peptide 920 is a potent inhibitor of E. coli LpxA (Ki = 50 nM). It is competitive with respect to acyl-ACP but not UDP-GlcNAc. The compact beta-turn structure of peptide 920 bound to LpxA may open previously uncharacterized approaches to the rational design of LpxA inhibitors with antibiotic activity.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, Box 3711 DUMC, Durham, NC 27710, USA.