2AIZ
Solution structure of peptidoglycan associated lipoprotein from Haemophilus influenza bound to UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine
- PDB DOI: https://doi.org/10.2210/pdb2AIZ/pdb
- BMRB: 6856
- BMRB: 6858
- BMRB: 6465
- Classification: MEMBRANE PROTEIN
- Organism(s): Haemophilus influenzae, synthetic construct
- Expression System: Escherichia coli BL21(DE3)
- Mutation(s): No
- Deposited: 2005-08-01 Released: 2006-03-14
Experimental Data Snapshot
- Method: SOLUTION NMR
- Conformers Calculated: 100
- Conformers Submitted: 20
- Selection Criteria: structures with the least restraint violations
wwPDB Validation 3D Report Full Report
This is version 2.0 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer membrane protein P6 (Fragment) | A [auth P] | 134 | Haemophilus influenzae | Mutation(s): 0 Gene Names: ompP6 | 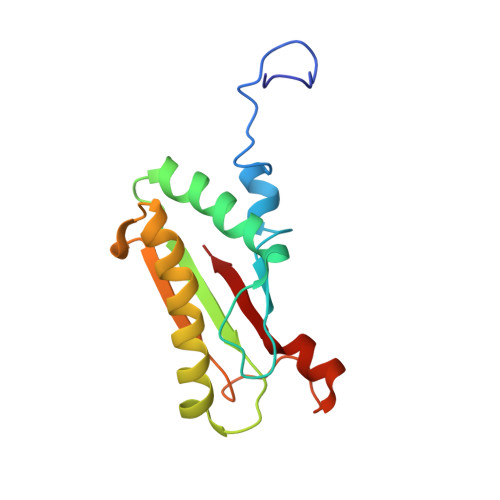 |
UniProt | |||||
Find proteins for P10324 (Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd)) Explore P10324 Go to UniProtKB: P10324 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10324 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | B [auth U] | 5 | synthetic construct | Mutation(s): 0 | 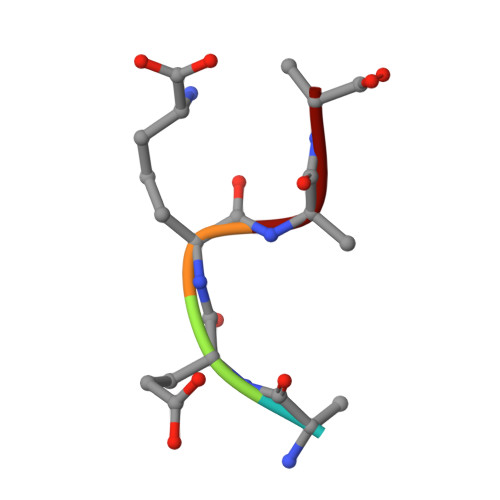 |
Sequence AnnotationsExpand | |||||
| |||||
Small Molecules
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| UDP Query on UDP | D [auth U] | URIDINE-5'-DIPHOSPHATE C9 H14 N2 O12 P2 XCCTYIAWTASOJW-XVFCMESISA-N |  | ||
| AMU Query on AMU | C [auth U] | N-acetyl-beta-muramic acid C11 H19 N O8 MNLRQHMNZILYPY-YVNCZSHWSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 6CL Query on 6CL | B [auth U] | L-PEPTIDE LINKING | C7 H15 N2 O4 |  | LYS |
Experimental Data & Validation
Experimental Data
- Method: SOLUTION NMR
- Conformers Calculated: 100
- Conformers Submitted: 20
- Selection Criteria: structures with the least restraint violations
Entry History
Deposition Data
- Released Date: 2006-03-14 Deposition Author(s): Parsons, L.M., Lin, F., Orban, J., Structure 2 Function Project (S2F)
Revision History (Full details and data files)
- Version 1.0: 2006-03-14
Type: Initial release - Version 1.1: 2008-04-30
Changes: Version format compliance - Version 1.2: 2011-07-13
Changes: Version format compliance - Version 1.3: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Data collection, Derived calculations, Structure summary - Version 2.0: 2023-02-15
Changes: Atomic model, Data collection, Database references, Derived calculations, Polymer sequence, Source and taxonomy, Structure summary













