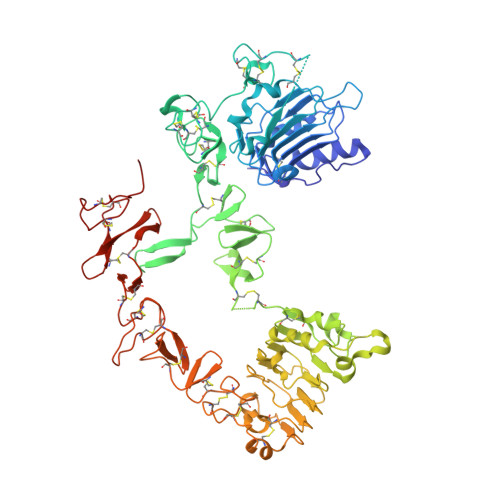
The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand
Bouyain, S., Longo, P.A., Li, S., Ferguson, K.M., Leahy, D.J.(2005) Proc Natl Acad Sci U S A 102: 15024-15029
- PubMed: 16203964
- DOI: https://doi.org/10.1073/pnas.0507591102
- Primary Citation of Related Structures:
2AHX - PubMed Abstract:
The human ErbB family of receptor tyrosine kinases comprises the epidermal growth factor receptor (EGFR/ErbB1/HER1), ErbB2 (HER2/Neu), ErbB3 (HER3), and ErbB4 (HER4). ErbBs play fundamental roles in cell growth and differentiation events in embryonic and adult tissues, and inappropriate ErbB activity has been implicated in several human cancers. We report here the 2.4 A crystal structure of the extracellular region of human ErbB4 in the absence of ligand and show that it adopts a tethered conformation similar to inactive forms of ErbB1 and ErbB3. This structure completes the gallery of unliganded ErbB receptors and demonstrates that all human ligand-binding ErbBs adopt the autoinhibited conformation. We also show that the binding of neuregulin-1beta to ErbB4 and ErbB3 and the binding of betacellulin to both ErbB4 and ErbB1 does not decrease at low pH, unlike the binding of epidermal growth factor and transforming growth factor-alpha to ErbB1. These results indicate an important role for ligand in determining pH-dependent binding and may explain different responses observed when the same ErbB receptor is stimulated by different ligands.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.