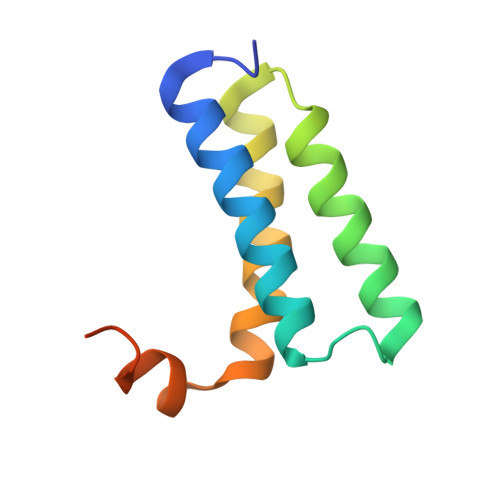
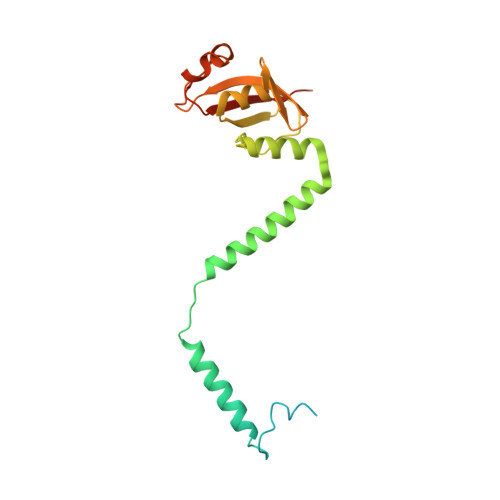
Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer
Zhai, Y.J., Sun, F., Li, X., Pang, H., Xu, X., Bartlam, M., Rao, Z.(2005) Nat Struct Mol Biol 12: 980-986
- PubMed: 16228002
- DOI: https://doi.org/10.1038/nsmb999
- Primary Citation of Related Structures:
2AHM - PubMed Abstract:
Coronavirus replication and transcription machinery involves multiple virus-encoded nonstructural proteins (nsp). We report the crystal structure of the hexadecameric nsp7-nsp8 supercomplex from the severe acute respiratory syndrome coronavirus at 2.4-angstroms resolution. nsp8 has a novel 'golf-club' fold with two conformations. The supercomplex is a unique hollow, cylinder-like structure assembled from eight copies of nsp8 and held tightly together by eight copies of nsp7. With an internal diameter of approximately 30 angstroms, the central channel has dimensions and positive electrostatic properties favorable for nucleic acid binding, implying that its role is to confer processivity on RNA-dependent RNA polymerase.
Organizational Affiliation:
Tsinghua-lnstitute of Biophysics Joint Research Group for Structural Biology, Tsinghua University, Beijing 100084, China.