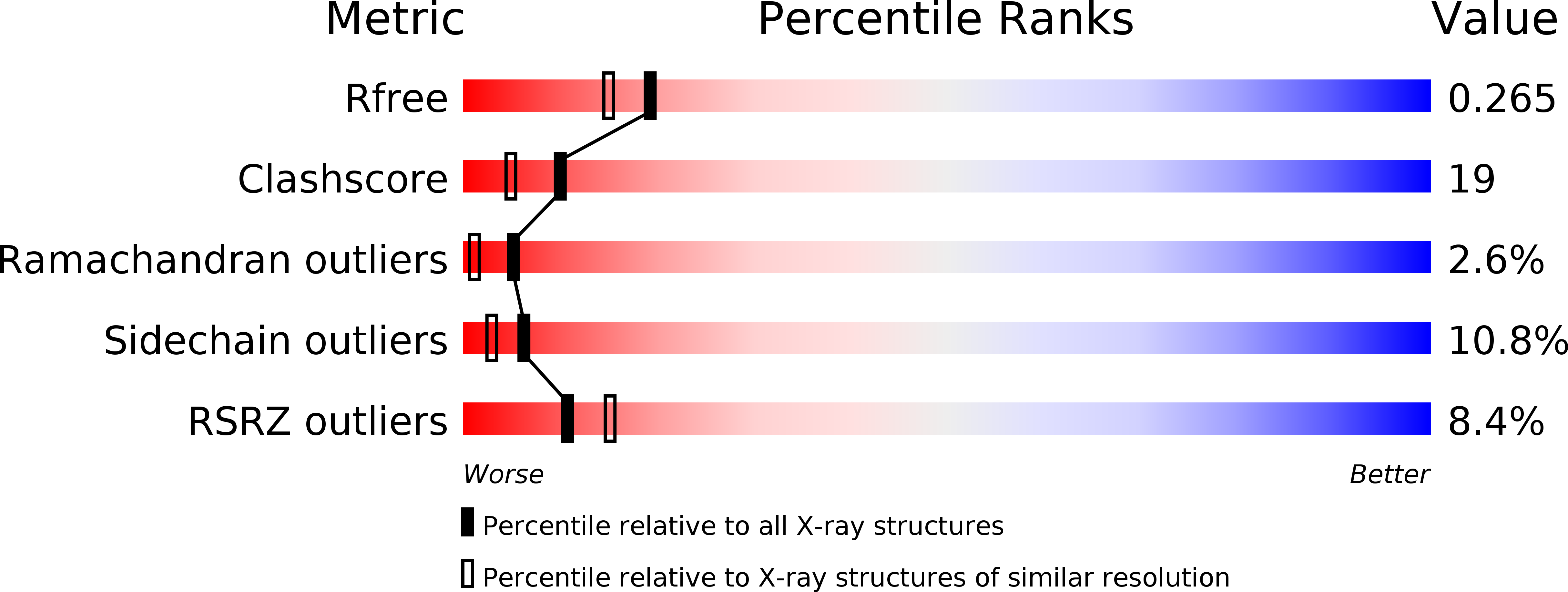
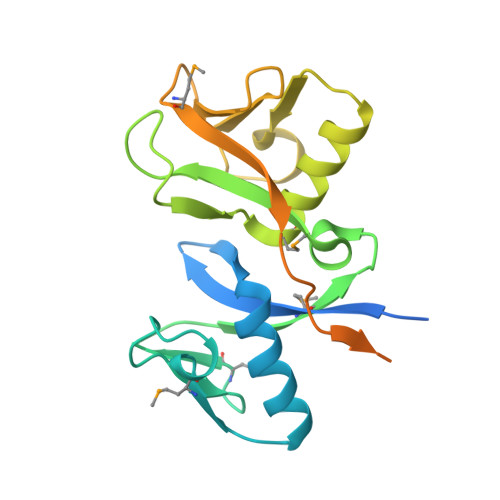
Structure and Notch Receptor Binding of the Tandem WWE Domain of Deltex.
Zweifel, M.E., Leahy, D.J., Barrick, D.(2005) Structure 13: 1599-1611
- PubMed: 16271883
- DOI: https://doi.org/10.1016/j.str.2005.07.015
- Primary Citation of Related Structures:
2A90 - PubMed Abstract:
Deltex is a cytosolic effector of Notch signaling thought to bind through its N-terminal domain to the Notch receptor. Here we report the structure of the Drosophila Deltex N-terminal domain, which contains two tandem WWE sequence repeats. The WWE repeats, which adopt a novel fold, are related by an approximate two-fold axis of rotation. Although the WWE repeats are structurally distinct, they interact extensively and form a deep cleft at their junction that appears well suited for ligand binding. The two repeats are thermodynamically coupled; this coupling is mediated in part by a conserved segment that is immediately C-terminal to the second WWE domain. We demonstrate that although the Deltex WWE tandem is monomeric in solution, it forms a heterodimer with the ankyrin domain of the Notch receptor. These results provide structural and functional insight into how Deltex modulates Notch signaling, and how WWE modules recognize targets for ubiquitination.
Organizational Affiliation:
T.C. Jenkins Department of Biophysics, Johns Hopkins University, Baltimore, Maryland 21218, USA.