Molecular dynamism of Fe-S cluster biosynthesis implicated by the structure of SufC(2)-SufD(2) complex
Wada, K., Sumi, N., Nagai, R., Iwasaki, K., Sato, T., Suzuki, K., Hasegawa, Y., Kitaoka, S., Minami, Y., Outten, F.W., Takahashi, Y., Fukuyama, K.(2009) J Mol Biol 387: 245-258
- PubMed: 19361433
- DOI: https://doi.org/10.1016/j.jmb.2009.01.054
- Primary Citation of Related Structures:
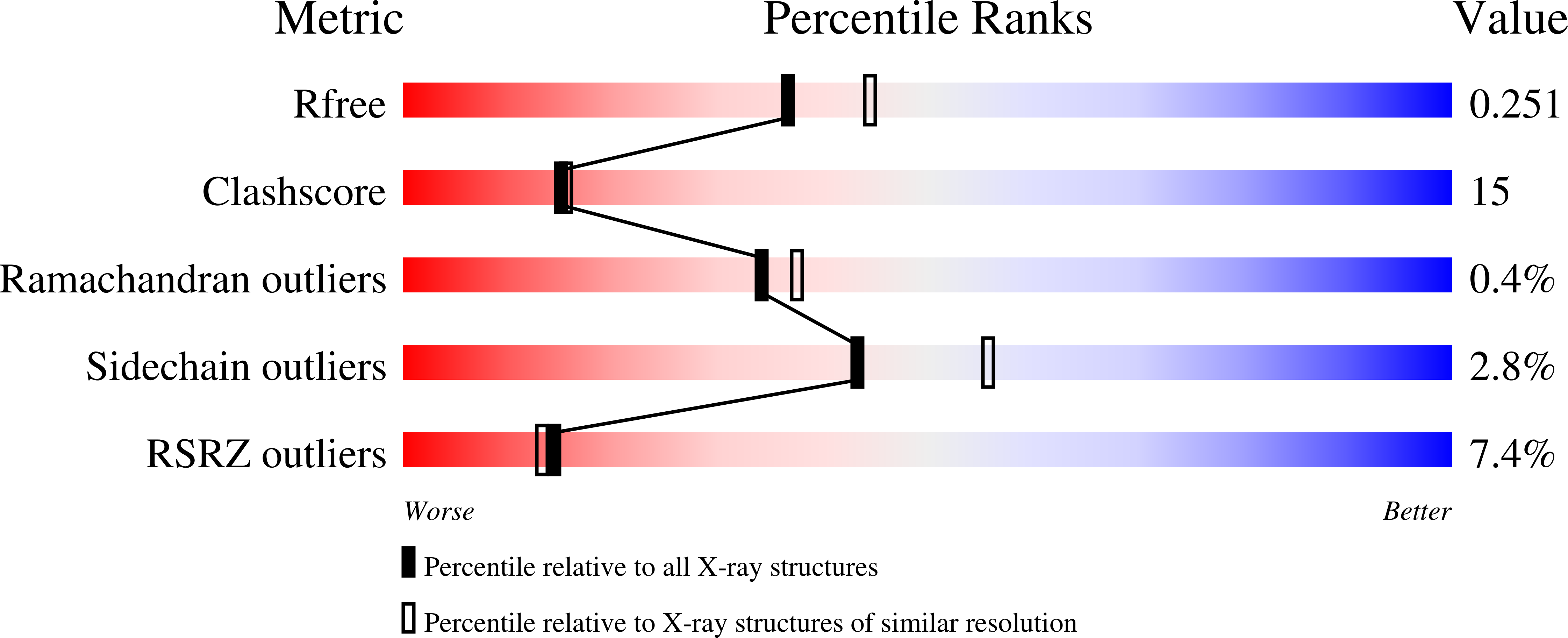
2ZU0 - PubMed Abstract:
Maturation of iron-sulfur (Fe-S) proteins is achieved by the SUF machinery in a wide number of eubacteria and archaea, as well as eukaryotic chloroplasts. This machinery is encoded in Escherichia coli by the sufABCDSE operon, where three Suf components, SufB, SufC, and SufD, form a complex and appear to provide an intermediary site for the Fe-S cluster assembly. Here, we report the quaternary structure of the SufC(2)-SufD(2) complex in which SufC is bound to the C-terminal domain of SufD. Comparison with the monomeric structure of SufC revealed conformational change of the active-site residues: SufC becomes competent for ATP binding and hydrolysis upon association with SufD. The two SufC subunits were spatially separated in the SufC(2)-SufD(2) complex, whereas cross-linking experiments in solution have indicated that two SufC molecules associate with each other in the presence of Mg(2+) and ATP. Such dimer formation of SufC may lead to a gross structural change of the SufC(2)-SufD(2) complex. Furthermore, genetic analysis of SufD revealed an essential histidine residue buried inside the dimer interface, suggesting that conformational change may expose this crucial residue. These findings, together with biochemical characterization of the SufB-SufC-SufD complex, have led us to propose a model for the Fe-S cluster biosynthesis in the complex.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, Osaka University, Toyonaka, Osaka, Japan.