Structural basis for dsRNA recognition by NS1 protein of influenza A virus
Cheng, A., Wong, S.M., Yuan, Y.A.(2009) Cell Res 19: 187-195
- PubMed: 18813227
- DOI: https://doi.org/10.1038/cr.2008.288
- Primary Citation of Related Structures:
2ZKO - PubMed Abstract:
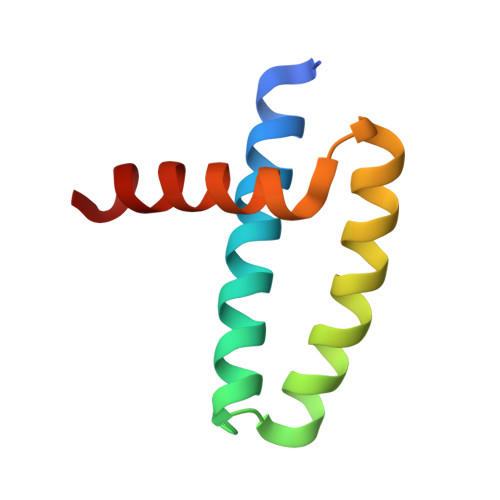
Influenza A viruses are important human pathogens causing periodic pandemic threats. Nonstructural protein 1 (NS1) protein of influenza A virus (NS1A) shields the virus against host defense. Here, we report the crystal structure of NS1A RNA-binding domain (RBD) bound to a double-stranded RNA (dsRNA) at 1.7A. NS1A RBD forms a homodimer to recognize the major groove of A-form dsRNA in a length-independent mode by its conserved concave surface formed by dimeric anti-parallel alpha-helices. dsRNA is anchored by a pair of invariable arginines (Arg38) from both monomers by extensive hydrogen bonds. In accordance with the structural observation, isothermal titration calorimetry assay shows that the unique Arg38-Arg38 pair and two Arg35-Arg46 pairs are crucial for dsRNA binding, and that Ser42 and Thr49 are also important for dsRNA binding. Agrobacterium co-infiltration assay further supports that the unique Arg38 pair plays important roles in dsRNA binding in vivo.Cell Research (2009) 19:187-195. doi: 10.1038/cr.2008.288; published online 23 September 2008.
Organizational Affiliation:
Genome and Structural Biology Program, Temasek Life Sciences Laboratory, National University of Singapore, 1 Research Link, Singapore 117604, Singapore.