Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Lee, E.F., Smith, B.J., Horne, W.S., Mayer, K.N., Evangelista, M., Colman, P.M., Gellman, S.H., Fairlie, W.D.(2011) Chembiochem 12: 2025
- PubMed: 21744457
- DOI: https://doi.org/10.1002/cbic.201100314
- Primary Citation of Related Structures:
2YJ1 - PubMed Abstract:
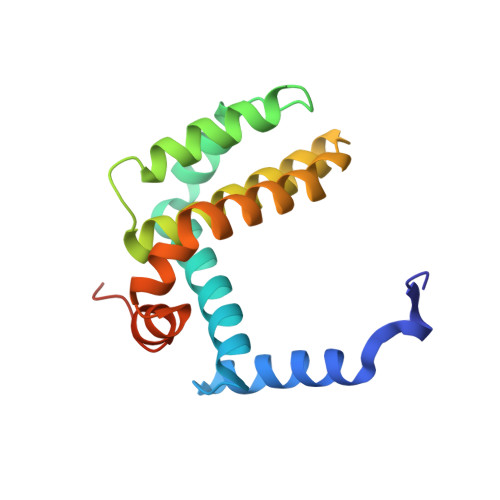
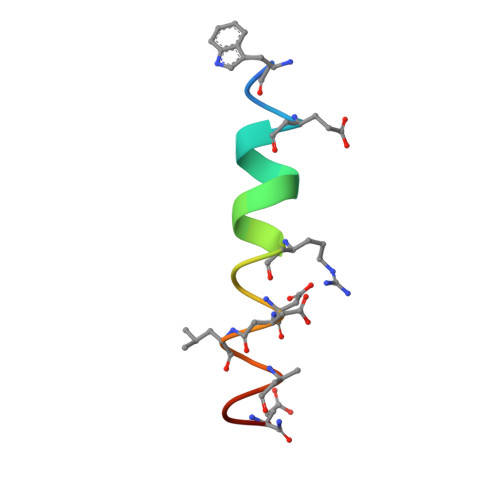
The crystal structure of a complex between the prosurvival protein Bcl-x(L) and an α/β-peptide 21-mer is described. The α/β-peptide contains six β-amino acid residues distributed periodically throughout the sequence and adopts an α-helix-like conformation that mimics the bioactive shape of the Puma BH3 domain. The α/β-peptide forms all of the noncovalent contacts that have previously been identified as necessary for recognition of the prosurvival protein by an authentic BH3 domain. Comparison of our α/β-peptide:Bcl-x(L) structure with structures of complexes between native BH3 domains and Bcl-2 family proteins reveals how subtle adjustments, including variations in helix radius and helix bowing, allow the α/β-peptide to engage Bcl-x(L) with high affinity. Geometric comparisons of the BH3-mimetic α/β-peptide with α/β-peptides in helix-bundle assemblies provide insight on the conformational plasticity of backbones that contain combinations of α- and β-amino acid residues. The BH3-mimetic α/β-peptide displays prosurvival protein-binding preferences distinct from those of Puma BH3 itself, even though these two oligomers have identical side-chain sequences. Our results suggest origins for this backbone-dependent change in selectivity.
Organizational Affiliation:
Structural Biology Division, The Walter and Eliza Hall Institute for Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia.