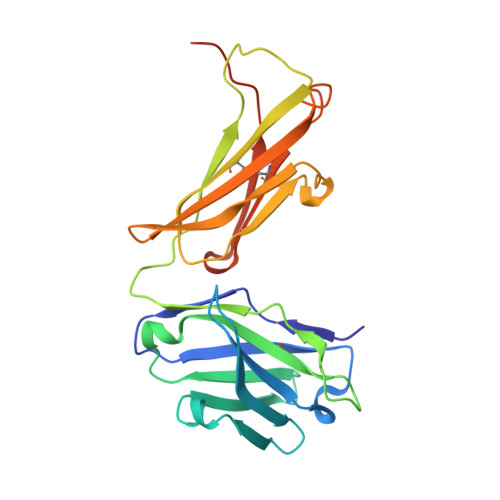
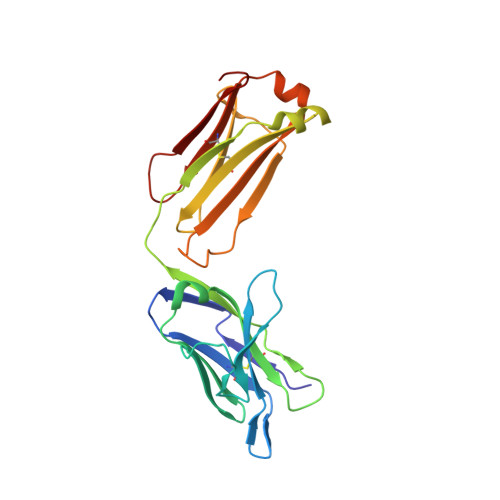
Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Dobritzsch, D., Lindh, I., Uysal, H., Nandakumar, K.S., Burkhardt, H., Schneider, G., Holmdahl, R.(2011) Arthritis Rheum 63: 3740
- PubMed: 22127694
- DOI: https://doi.org/10.1002/art.30611
- Primary Citation of Related Structures:
2Y5T - PubMed Abstract:
In rheumatoid arthritis, joint inflammation and cartilage destruction are mediated by autoantibodies directed to various self antigens. Type II collagen (CII)-specific antibodies are likely to play a role in this process and have been shown to induce experimental arthritis in susceptible animals. The purpose of this study was to reveal how arthritogenic autoantibodies recognize native CII in its triple-helical conformation.
Organizational Affiliation:
Karolinska Institute, Stockholm, Sweden.