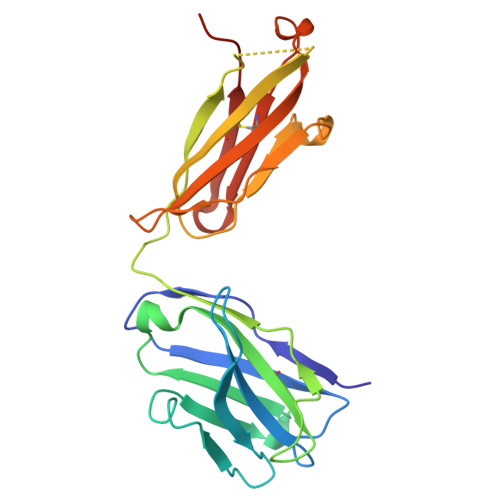
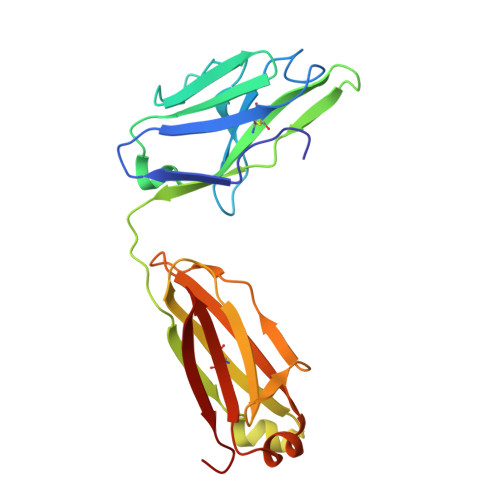
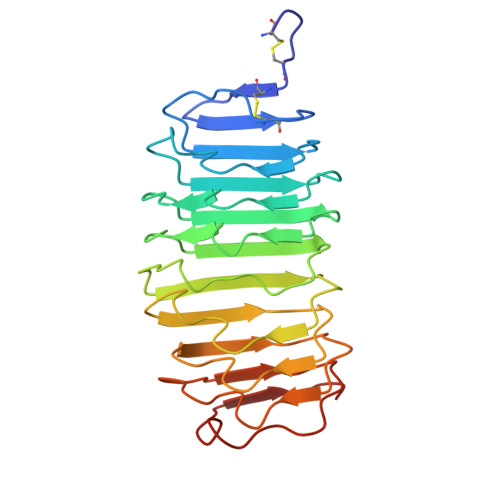
Crystal Structure of the Tsh Receptor (Tshr) Bound to a Blocking-Type Tshr Autoantibody.
Sanders, P., Young, S., Sanders, J., Kabelis, K., Baker, S., Sullivan, A., Evans, M., Clark, J., Wilmot, J., Hu, X., Roberts, E., Powell, M., Nunez Miguel, R., Furmaniak, J., Rees Smith, B.(2011) J Mol Endocrinol 46: 81
- PubMed: 21247981
- DOI: https://doi.org/10.1530/JME-10-0127
- Primary Citation of Related Structures:
2XWT - PubMed Abstract:
A complex of the TSH receptor extracellular domain (amino acids 22-260; TSHR260) bound to a blocking-type human monoclonal autoantibody (K1-70) was purified, crystallised and the structure solved at 1.9 Å resolution. K1-70 Fab binds to the concave surface of the TSHR leucine-rich domain (LRD) forming a large interface (2565 Å(2)) with an extensive network of ionic, polar and hydrophobic interactions. Mutation of TSHR or K1-70 residues showing strong interactions in the solved structure influenced the activity of K1-70, indicating that the binding detail observed in the complex reflects interactions of K1-70 with intact, functionally active TSHR. Unbound K1-70 Fab was prepared and crystallised to 2.22 Å resolution. Virtually no movement was observed in the atoms of K1-70 residues on the binding interface compared with unbound K1-70, consistent with 'lock and key' binding. The binding arrangements in the TSHR260-K1-70 Fab complex are similar to previously observed for the TSHR260-M22 Fab complex; however, K1-70 clasps the concave surface of the TSHR LRD in approximately the opposite orientation (rotated 155°) to M22. The blocking autoantibody K1-70 binds more N-terminally on the TSHR concave surface than either the stimulating autoantibody M22 or the hormone TSH, and this may reflect its different functional activity. The structure of TSHR260 in the TSHR260-K1-70 and TSHR260-M22 complexes show a root mean square deviation on all C(α) atoms of only 0.51 Å. These high-resolution crystal structures provide a foundation for developing new strategies to understand and control TSHR activation and the autoimmune response to the TSHR.
Organizational Affiliation:
FIRS Laboratories, RSR Ltd, Parc Ty Glas, Llanishen, Cardiff CF14 5DU, UK.