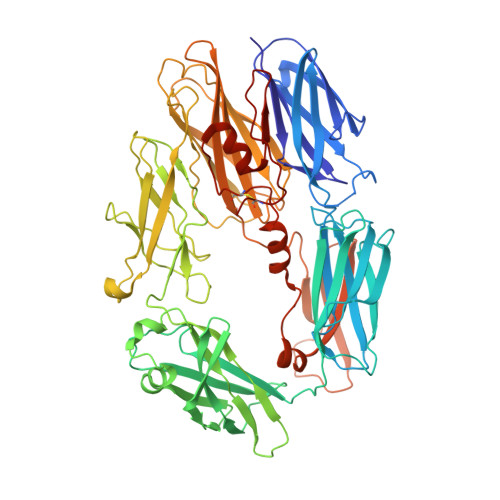
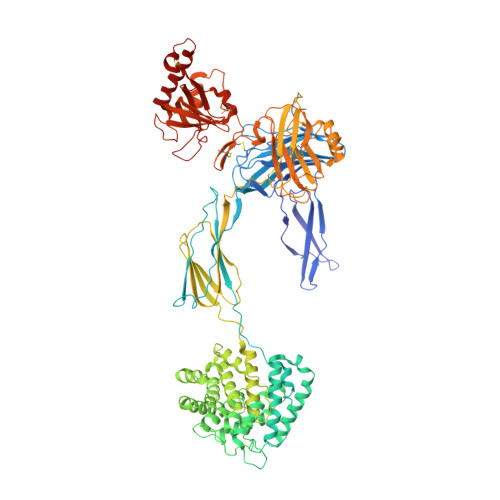
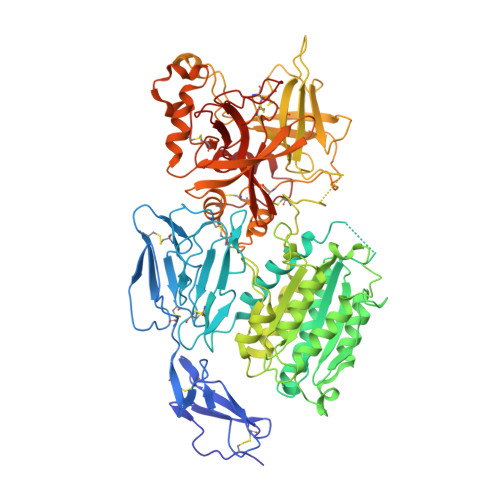
Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Forneris, F., Ricklin, D., Wu, J., Tzekou, A., Wallace, R.S., Lambris, J.D., Gros, P.(2010) Science 330: 1816
- PubMed: 21205667
- DOI: https://doi.org/10.1126/science.1195821
- Primary Citation of Related Structures:
2XW9, 2XWA, 2XWB, 2XWJ - PubMed Abstract:
Activation of the complement cascade induces inflammatory responses and marks cells for immune clearance. In the central complement-amplification step, a complex consisting of surface-bound C3b and factor B is cleaved by factor D to generate active convertases on targeted surfaces. We present crystal structures of the pro-convertase C3bB at 4 angstrom resolution and its complex with factor D at 3.5 angstrom resolution. Our data show how factor B binding to C3b forms an open "activation" state of C3bB. Factor D specifically binds the open conformation of factor B through a site distant from the catalytic center and is activated by the substrate, which displaces factor D's self-inhibitory loop. This concerted proteolytic mechanism, which is cofactor-dependent and substrate-induced, restricts complement amplification to C3b-tagged target cells.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH Utrecht, Netherlands.