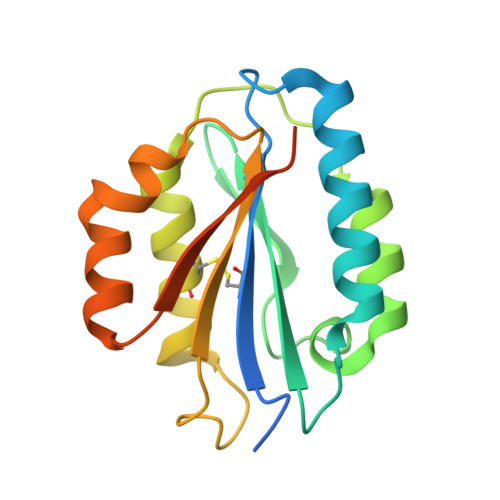
Structure of the Micronemal Protein 2 (Mic2) A/I Domain from Toxoplasma Gondii.
Tonkin, M.L., Grujic, O., Pearce, M., Crawford, J., Boulanger, M.J.(2010) Protein Sci 19: 1985
- PubMed: 20684023
- DOI: https://doi.org/10.1002/pro.477
- Primary Citation of Related Structures:
2XGG - PubMed Abstract:
Toxoplasma gondii is a widespread zoonotic pathogen capable of causing serious disease in humans and animals. As an obligate intracellular parasite, T. gondii relies on the orchestrated secretion of proteins from its apical complex organelles including the multimodular, transmembrane micronemal protein 2 (MIC2) that couples recognition of the host cell with cytoskeletal reorganization of the parasite to drive invasion. To probe the basis by which the von Willebrand Factor A (vWA)-Integrin like module of TgMIC2 engages the host cell, we solved the crystal structure of a truncated form of TgMIC2A/I (TgMIC2A/Ic) phased by iodide SIRAS and refined to a resolution of 2.05 Å. The TgMIC2A/Ic core is organized into a central twisted beta sheet flanked by α-helices consistent with a canonical vWA fold. A restricted basic patch serves as the putative heparin binding site, but no heparin binding was detected in native gel shift assays. Furthermore, no metal was observed in the metal ion dependent adhesion site (MIDAS). Structural overlays with homologous A/I domains reveal a divergent organization of the MIDAS β4-α4 loop in TgMIC2A/Ic, which is stabilized through the burial of Phe195 into a deep pocket formed by Gly185. Intriguingly, Gly185 appears to be unique among A/I domains to TgMIC2A/I suggesting that the divergent loop conformation may also be unique to TgMIC2A/I. Although lacking the C-terminal extension, the TgMIC2A/Ic structure reported here is the first of an A/I domain from an apicomplexan parasite and provides valuable insight into defining the molecular recognition of host cells by these widespread pathogens.
Organizational Affiliation:
Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia, Canada.