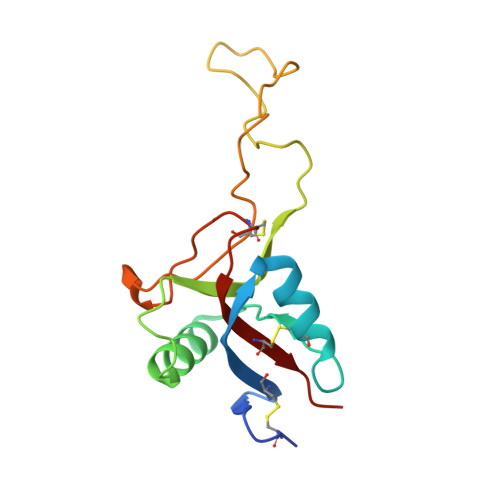
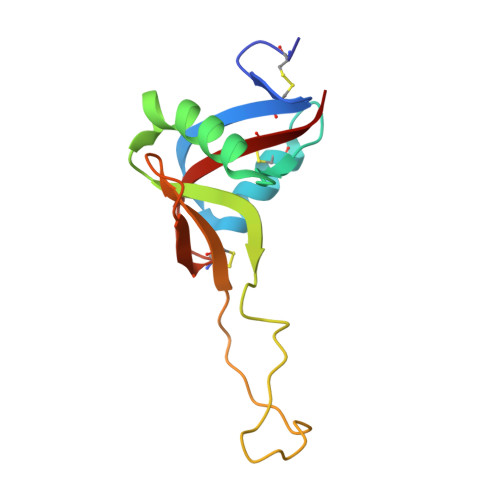
Crystal Structure of Rhodocytin, a Ligand for the Platelet-Activating Receptor Clec-2.
Watson, A.A., Eble, J.A., O'Callaghan, C.A.(2008) Protein Sci 17: 1611
- PubMed: 18583525
- DOI: https://doi.org/10.1110/ps.035568.108
- Primary Citation of Related Structures:
2VRP - PubMed Abstract:
Binding of the snake venom protein rhodocytin to CLEC-2, a receptor on the surface of human platelets, initiates a signaling cascade leading to platelet activation and aggregation. We have previously solved the structure of CLEC-2. The 2.4 A resolution crystal structure of rhodocytin presented here demonstrates that it is the first snake venom or other C-type lectin-like protein to assemble as a non-disulfide linked (alphabeta)(2) tetramer. Rhodocytin is highly adapted for interaction with CLEC-2 and displays a concave binding surface, which is highly complementary to the experimentally determined binding interface on CLEC-2. Using computational dynamic methods, surface electrostatic charge and hydrophobicity analyses, and protein-protein docking predictions, we propose that the (alphabeta)(2) rhodocytin tetramer induces clustering of CLEC-2 receptors on the platelet surface, which will trigger major signaling events resulting in platelet activation and aggregation.
Organizational Affiliation:
Henry Wellcome Building for Molecular Physiology, University of Oxford, Oxford OX3 7BN, United Kingdom.