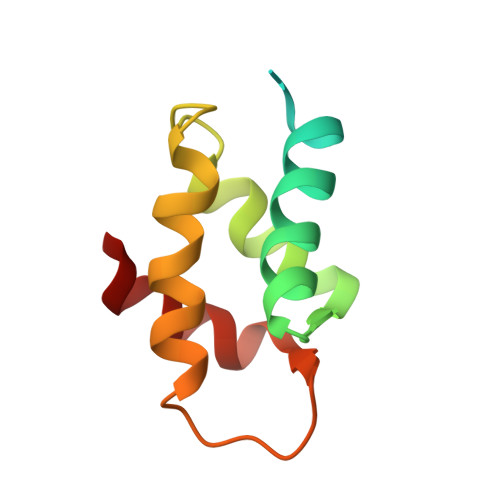
The Solution Structure of the Amino-Terminal Domain of Human DNA Polymerase Epsilon Subunit B is Homologous to C-Domains of Aaa+ Proteins.
Nuutinen, T., Tossavainen, H., Fredriksson, K., Pirila, P., Permi, P., Pospiech, H., Syvaoja, J.E.(2008) Nucleic Acids Res 36: 5102-5110
- PubMed: 18676977
- DOI: https://doi.org/10.1093/nar/gkn497
- Primary Citation of Related Structures:
2V6Z - PubMed Abstract:
DNA polymerases alpha, delta and epsilon are large multisubunit complexes that replicate the bulk of the DNA in the eukaryotic cell. In addition to the homologous catalytic subunits, these enzymes possess structurally related B subunits, characterized by a carboxyterminal calcineurin-like and an aminoproximal oligonucleotide/oligosaccharide binding-fold domain. The B subunits also share homology with the exonuclease subunit of archaeal DNA polymerases D. Here, we describe a novel domain specific to the N-terminus of the B subunit of eukaryotic DNA polymerases epsilon. The N-terminal domain of human DNA polymerases epsilon (Dpoe2NT) expressed in Escherichia coli was characterized. Circular dichroism studies demonstrated that Dpoe2NT forms a stable, predominantly alpha-helical structure. The solution structure of Dpoe2NT revealed a domain that consists of a left-handed superhelical bundle. Four helices are arranged in two hairpins and the connecting loops contain short beta-strand segments that form a short parallel sheet. DALI searches demonstrated a striking structural similarity of the Dpoe2NT with the alpha-helical subdomains of ATPase associated with various cellular activity (AAA+) proteins (the C-domain). Like C-domains, Dpoe2NT is rich in charged amino acids. The biased distribution of the charged residues is reflected by a polarization and a considerable dipole moment across the Dpoe2NT. Dpoe2NT represents the first C-domain fold not associated with an AAA+ protein.
Organizational Affiliation:
Faculty of Biosciences, University of Joensuu, Finland.