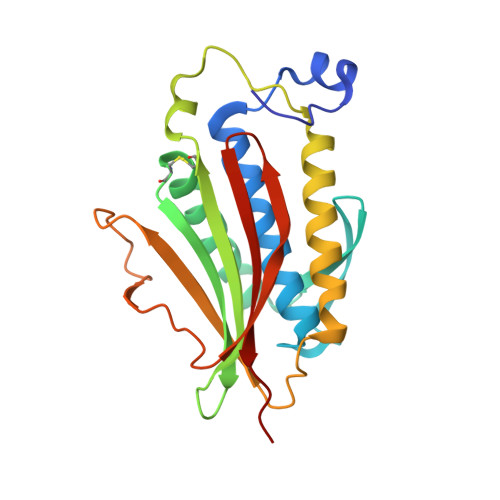
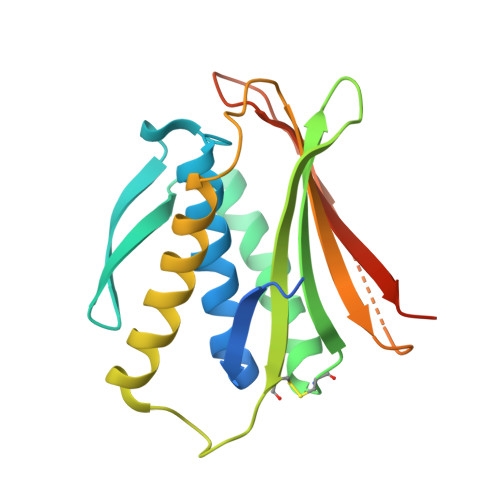
The MAD2 Conformational Dimer: Structure and Implications for the Spindle Assembly Checkpoint
Mapelli, M., Massimiliano, L., Santaguida, S., Musacchio, A.(2007) Cell 131: 730
- PubMed: 18022367
- DOI: https://doi.org/10.1016/j.cell.2007.08.049
- Primary Citation of Related Structures:
2V64 - PubMed Abstract:
The 25 kDa Mad2 protein is a key player in the spindle assembly checkpoint, a safeguard against chromosome segregation errors in mitosis. Mad2 combines three unusual properties. First, Mad2 adopts two conformations with distinct topologies, open (O) and closed (C) Mad2. Second, C-Mad2 forms topological links with its two best-characterized protein ligands, Mad1 and Cdc20. Third, O-Mad2 and C-Mad2 engage in a "conformational" dimer that is essential for spindle checkpoint function in different organisms. The crystal structure of the O-Mad2-C-Mad2 conformational dimer, reported here, reveals an asymmetric interface that explains the selective dimerization of the O-Mad2 and C-Mad2 conformers. The structure also identifies several buried hydrophobic residues whose rearrangement correlates with the Mad2 topological change. The structure of the O-Mad2-C-Mad2 conformational dimer is consistent with a catalytic model in which a C-Mad2 template facilitates the binding of O-Mad2 to Cdc20, the target of Mad2 in the spindle checkpoint.
Organizational Affiliation:
Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, I-20139, Milan, Italy. marina.mapelli@ifom-ieo-campus.it