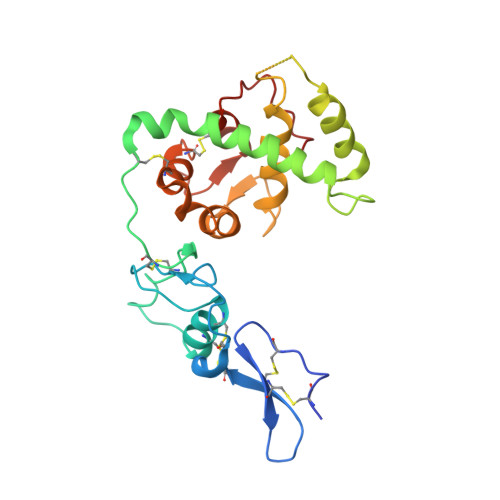
Structural Basis of Sequence-Specific Collagen Recognition by Sparc.
Hohenester, E., Sasaki, T., Giudici, C., Farndale, R.W., Bachinger, H.P.(2008) Proc Natl Acad Sci U S A 105: 18273
- PubMed: 19011090
- DOI: https://doi.org/10.1073/pnas.0808452105
- Primary Citation of Related Structures:
2V53 - PubMed Abstract:
Protein interactions with the collagen triple helix play a critical role in collagen fibril formation, cell adhesion, and signaling. However, structural insight into sequence-specific collagen recognition is limited to an integrin-peptide complex. A GVMGFO motif in fibrillar collagens (O denotes 4-hydroxyproline) binds 3 unrelated proteins: von Willebrand factor (VWF), discoidin domain receptor 2 (DDR2), and the extracellular matrix protein SPARC/osteonectin/BM-40. We report the crystal structure at 3.2 A resolution of human SPARC bound to a triple-helical 33-residue peptide harboring the promiscuous GVMGFO motif. SPARC recognizes the GVMGFO motifs of the middle and trailing collagen chains, burying a total of 720 A(2) of solvent-accessible collagen surface. SPARC binding does not distort the canonical triple helix of the collagen peptide. In contrast, a critical loop in SPARC is substantially remodelled upon collagen binding, creating a deep pocket that accommodates the phenylalanine residue of the trailing collagen chain ("Phe pocket"). This highly restrictive specificity pocket is shared with the collagen-binding integrin I-domains but differs strikingly from the shallow collagen-binding grooves of the platelet receptor glycoprotein VI and microbial adhesins. We speculate that binding of the GVMGFO motif to VWF and DDR2 also results in structural changes and the formation of a Phe pocket.
Organizational Affiliation:
Department of Life Sciences, Imperial College London, London SW7 2AZ, United Kingdom. e.hohenester@imperial.ac.uk