The structure of the Haemophilus influenzae HMW1 pro-piece reveals a structural domain essential for bacterial two-partner secretion.
Yeo, H.J., Yokoyama, T., Walkiewicz, K., Kim, Y., Grass, S., Geme, J.W.(2007) J Biol Chem 282: 31076-31084
- PubMed: 17699157
- DOI: https://doi.org/10.1074/jbc.M705750200
- Primary Citation of Related Structures:
2ODL - PubMed Abstract:
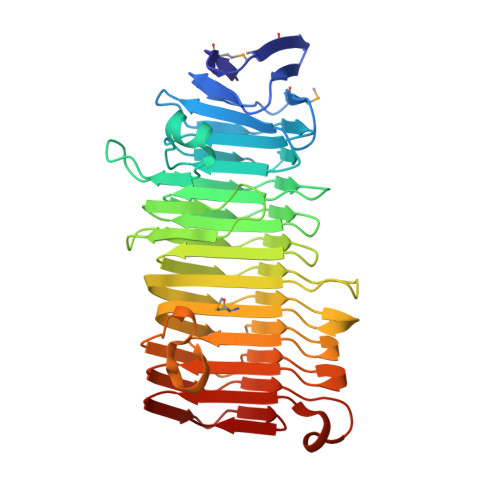
In pathogenic Gram-negative bacteria, many virulence factors are secreted via the two-partner secretion pathway, which consists of an exoprotein called TpsA and a cognate outer membrane translocator called TpsB. The HMW1 and HMW2 adhesins are major virulence factors in nontypeable Haemophilus influenzae and are prototype two-partner secretion pathway exoproteins. A key step in the delivery of HMW1 and HMW2 to the bacterial surface involves targeting to the HMW1B and HMW2B outer membrane translocators by an N-terminal region called the secretion domain. Here we present the crystal structure at 1.92 A of the HMW1 pro-piece (HMW1-PP), a region that contains the HMW1 secretion domain and is cleaved and released during HMW1 secretion. Structural analysis of HMW1-PP revealed a right-handed beta-helix fold containing 12 complete parallel coils and one large extra-helical domain. Comparison of HMW1-PP and the Bordetella pertussis FHA secretion domain (Fha30) reveals limited amino acid homology but shared structural features, suggesting that diverse TpsA proteins have a common structural domain required for targeting to cognate TpsB proteins. Further comparison of HMW1-PP and Fha30 structures may provide insights into the keen specificity of TpsA-TpsB interactions.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Houston, Houston, Texas 77204, USA. hyeo@uh.edu