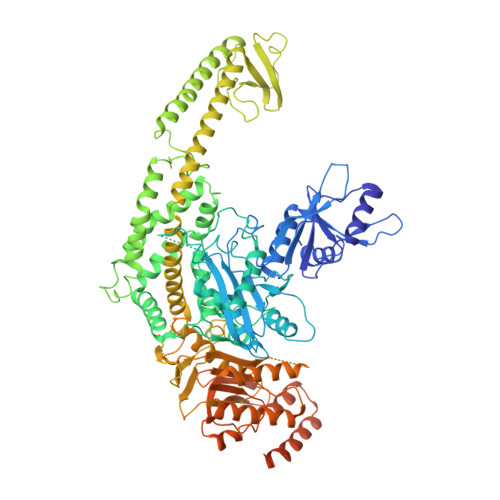
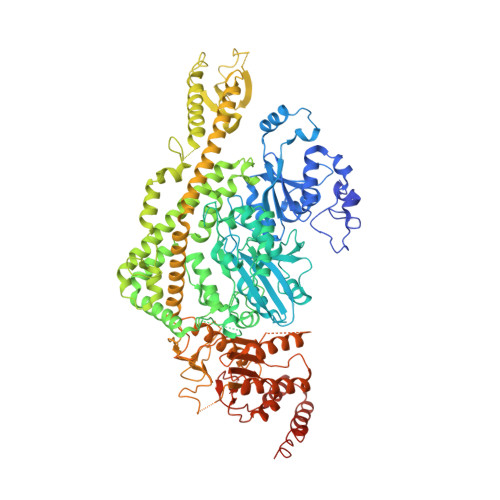
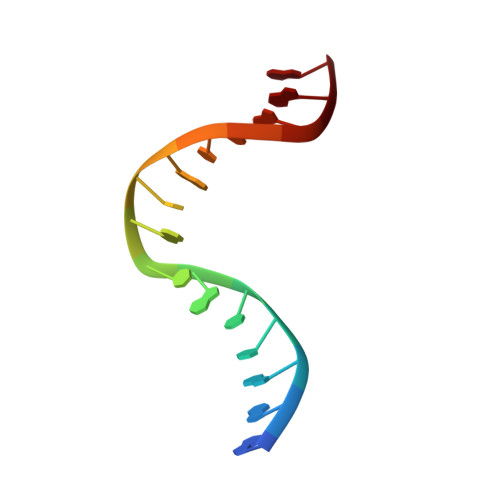
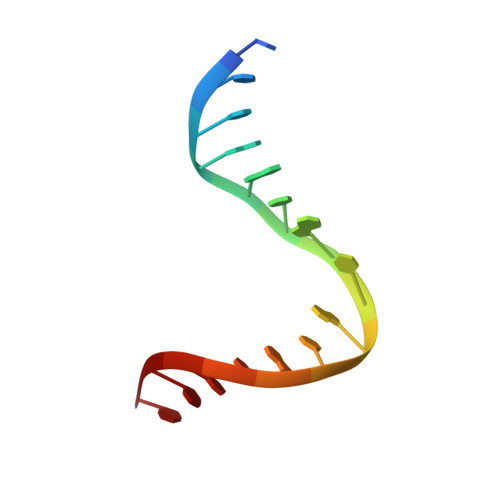
Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Warren, J.J., Pohlhaus, T.J., Changela, A., Iyer, R.R., Modrich, P.L., Beese, L.S.(2007) Mol Cell 26: 579-592
- PubMed: 17531815
- DOI: https://doi.org/10.1016/j.molcel.2007.04.018
- Primary Citation of Related Structures:
2O8B, 2O8C, 2O8D, 2O8E, 2O8F - PubMed Abstract:
Mismatch repair (MMR) ensures the fidelity of DNA replication, initiates the cellular response to certain classes of DNA damage, and has been implicated in the generation of immune diversity. Each of these functions depends on MutSalpha (MSH2*MSH6 heterodimer). Inactivation of this protein complex is responsible for tumor development in about half of known hereditary nonpolyposis colorectal cancer kindreds and also occurs in sporadic tumors in a variety of tissues. Here, we describe a series of crystal structures of human MutSalpha bound to different DNA substrates, each known to elicit one of the diverse biological responses of the MMR pathway. All lesions are recognized in a similar manner, indicating that diversity of MutSalpha-dependent responses to DNA lesions is generated in events downstream of this lesion recognition step. This study also allows rigorous mapping of cancer-causing mutations and furthermore suggests structural pathways for allosteric communication between different regions within the heterodimer.
Organizational Affiliation:
Department of Biochemistry, Duke University Medical Center, Durham, NC 27710, USA.