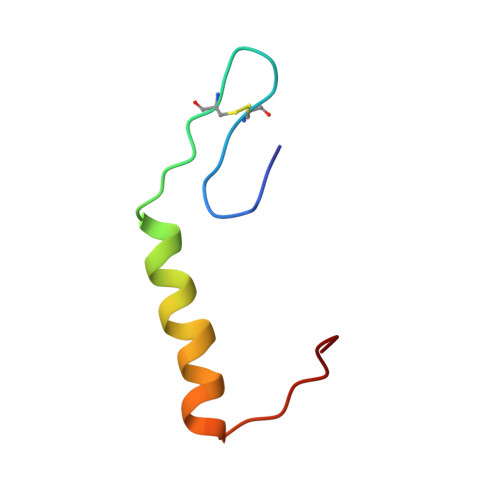
Solution Structure of Enterocin HF, an Antilisterial Bacteriocin Produced by Enterococcus faecium M3K31.
Arbulu, S., Lohans, C.T., van Belkum, M.J., Cintas, L.M., Herranz, C., Vederas, J.C., Hernandez, P.E.(2015) J Agric Food Chem 63: 10689-10695
- PubMed: 26585399
- DOI: https://doi.org/10.1021/acs.jafc.5b03882
- Primary Citation of Related Structures:
2N4K - PubMed Abstract:
The solution structure of enterocin HF (EntHF), a class IIa bacteriocin of 43 amino acids produced by Enterococcus faecium M3K31, was evaluated by CD and NMR spectroscopy. Purified EntHF was unstructured in water, but CD analysis supports that EntHF adopts an α-helical conformation when exposed to increasing concentrations of trifluoroethanol. Furthermore, NMR spectroscopy indicates that this bacteriocin adopts an antiparallel β-sheet structure in the N-terminal region (residues 1-17), followed by a well-defined central α-helix (residues 19-30) and a more disordered C-terminal end (residues 31-43). EntHF could be structurally organized into three flexible regions that might act in a coordinated manner. This is in agreement with the absence of long-range nuclear Overhauser effect signals between the β-sheet domain and the C-terminal end of the bacteriocin. The 3D structure recorded for EntHF fits emerging facts regarding target recognition and mode of action of class IIa bacteriocins.
Organizational Affiliation:
Departamento de Nutrición, Bromatologı́a y Tecnologı́a de los Alimentos, Facultad de Veterinaria, Universidad Complutense de Madrid (UCM) , Avenida Puerta de Hierro s/n, 28040 Madrid, Spain.