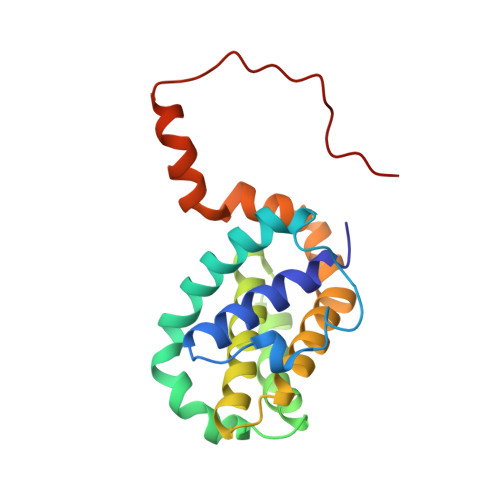
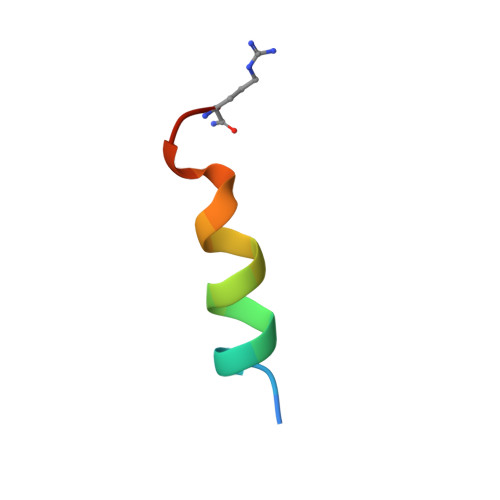
NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
Wysoczanski, P., Mart, R.J., Loveridge, E.J., Williams, C., Whittaker, S.B., Crump, M.P., Allemann, R.K.(2012) J Am Chem Soc 134: 7644-7647
- PubMed: 22515821
- DOI: https://doi.org/10.1021/ja302390a
- Primary Citation of Related Structures:
2LP8, 2LPC - PubMed Abstract:
The Bcl-2 family of proteins includes the major regulators and effectors of the intrinsic apoptosis pathway. Cancers are frequently formed when activation of the apoptosis mechanism is compromised either by misregulated expression of prosurvival family members or, more frequently, by damage to the regulatory pathways that trigger intrinsic apoptosis. Short peptides derived from the pro-apoptotic members of the Bcl-2 family can activate mechanisms that ultimately lead to cell death. The recent development of photocontrolled peptides that are able to change their conformation and activity upon irradiation with an external light source has provided new tools to target cells for apoptosis induction with temporal and spatial control. Here, we report the first NMR solution structure of a photoswitchable peptide derived from the proapoptotic protein Bak in complex with the antiapoptotic protein Bcl-x(L). This structure provides insight into the molecular mechanism, by which the increased affinity of such photopeptides compared to their native forms is achieved, and offers a rationale for the large differences in the binding affinities between the helical and nonhelical states.
Organizational Affiliation:
School of Chemistry and Cardiff Catalysis Institute, Cardiff University, Main Building, Park Place, Cardiff CF10 3AT, United Kingdom.