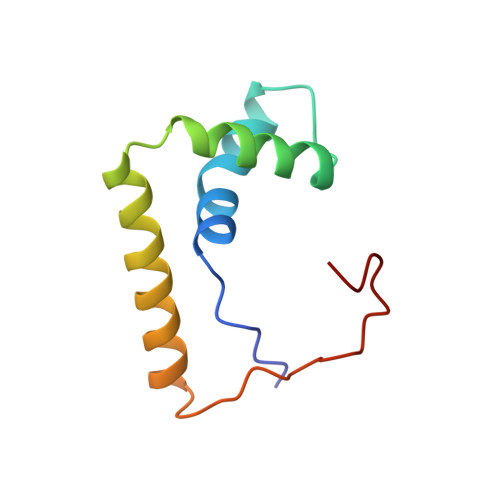
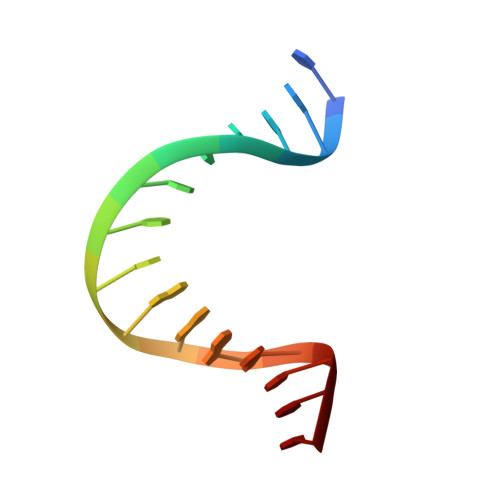
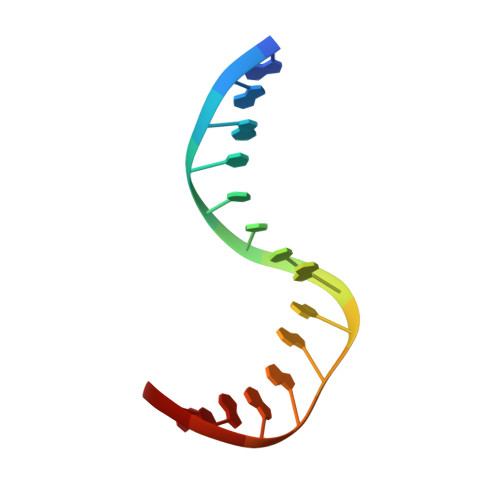
Structural basis for DNA bending by the architectural transcription factor LEF-1.
Love, J.J., Li, X., Case, D.A., Giese, K., Grosschedl, R., Wright, P.E.(1995) Nature 376: 791-795
- PubMed: 7651541
- DOI: https://doi.org/10.1038/376791a0
- Primary Citation of Related Structures:
2LEF - PubMed Abstract:
Lymphoid enhancer-binding factor (LEF-1) and the closely related T-cell factor 1 (TCF-1) are sequence-specific and cell-type-specific DNA-binding proteins that play important regulatory roles in organogenesis and thymocyte differentiation. LEF-1 participates in regulation of the enhancer associated with the T cell receptor (TCR)-alpha gene by inducing a sharp bend in the DNA and facilitating interactions between Ets-1, PEBP2-alpha, and ATF/CREB, transcription factors bound at sites flanking the LEF-1 site. It seems that LEF-1 plays an architectural role in the assembly and function of this regulatory nucleoprotein complex. LEF-1 recognizes a specific nucleotide sequence through a high-mobility-group (HMG) domain. Proteins containing HMG domains bind DNA in the minor groove, bend the double helix, and recognize four-way junctions and other irregular DNA structures. Here we report the solution structure of a complex of the LEF-1 HMG domain and adjacent basic region with its cognate DNA. The structure reveals the HMG domain bound in the widened minor groove of a markedly distorted and bent double helix. The basic region binds across the narrowed major groove and contributes to DNA recognition.
Organizational Affiliation:
Department of Molecular Biology, Scripps Research Institute, La Jolla, California 92037, USA.