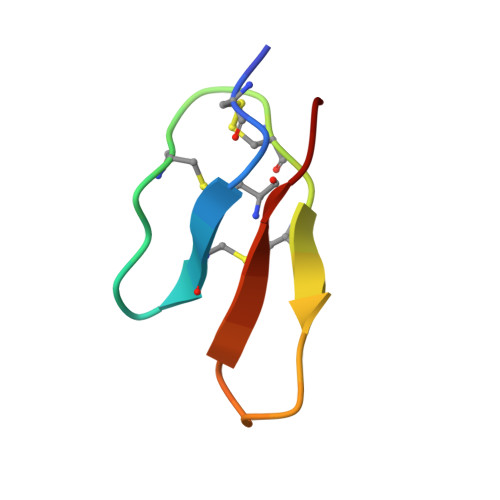
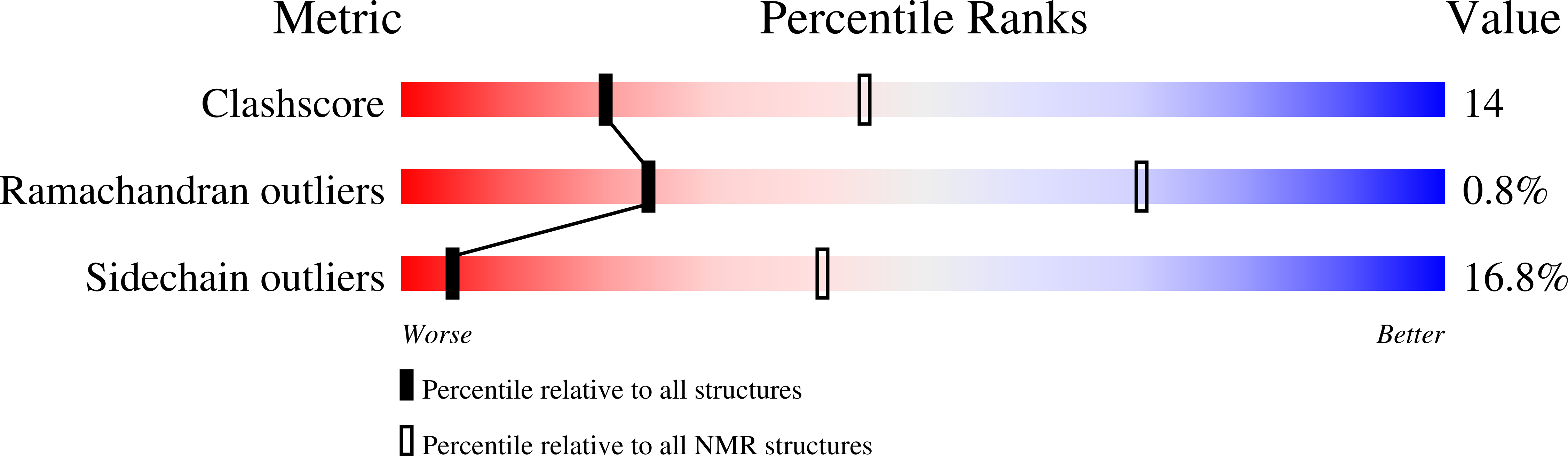Isolation and characterization of peptides from Momordica cochinchinensis seeds.
Chan, L.Y., Wang, C.K., Major, J.M., Greenwood, K.P., Lewis, R.J., Craik, D.J., Daly, N.L.(2009) J Nat Prod 72: 1453-1458
- PubMed: 19711988
- DOI: https://doi.org/10.1021/np900174n
- Primary Citation of Related Structures:
2KNP - PubMed Abstract:
The plant Momordica cochinchinensis has traditionally been used in Chinese medicine to treat a variety of illnesses. A range of bioactive molecules have been isolated from this plant, including peptides, which are the focus of this study. Here we report the isolation and characterization of two novel peptides, MCoCC-1 and MCoCC-2, containing 33 and 32 amino acids, respectively, which are toxic against three cancer cell lines. The two peptides are highly homologous to one another, but show no sequence similarity to known peptides. Elucidation of the three-dimensional structure of MCoCC-1 suggests the presence of a cystine knot motif, also found in a family of trypsin inhibitor peptides from this plant. However, unlike its structural counterparts, MCoCC-1 does not inhibit trypsin. MCoCC-1 has a well-defined structure, characterized mainly by a triple-stranded antiparallel beta-sheet, but unlike the majority of cystine knot proteins MCoCC-1 contains a disordered loop presumably as a result of flexibility in a localized region of the molecule. Of the cell lines tested, MCoCC-1 is the most toxic against a human melanoma cell line (MM96L) and is nonhemolytic to human erythrocytes. The role of these peptides within the plant remains to be determined.
Organizational Affiliation:
The University of Queensland, Institute for Molecular Bioscience, Brisbane QLD 4072, Australia.