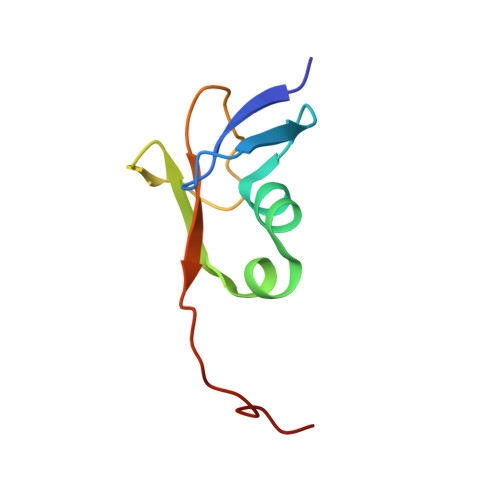
Solution NMR structure of the TGS domain of PG1808 from Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target PgR122A (418-481)
Yang, Y., Ramelot, T.A., Cort, J.R., Lee, D., Ciccosanti, C., Hamilton, K., Nair, R., Rost, B., Acton, T.B., Xiao, R., Swapna, G., Everett, J.K., Montelione, G.T., Kennedy, M.A.To be published.