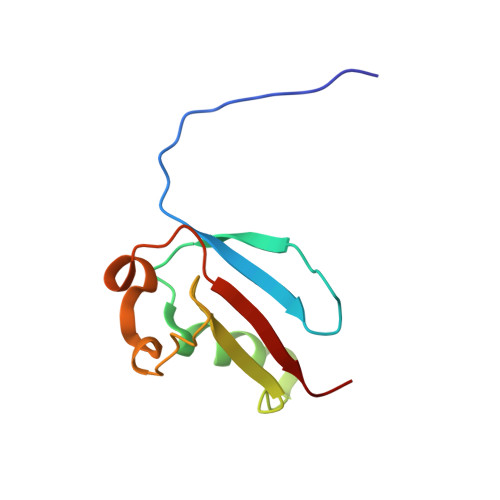
NMR Solution Structure of Tbulin folding Cofactor B obtained from Arabidopsis thaliana: Northeast
Mani, R., Swapna, G.V.T., Shastry, R., Foote, E., Ciccosanti, C., Jiang, M., Xiao, R., Nair, R., Everett, J., Huang, Y.J., Acton, T., Rost, B., Montelione, G.T.To be published.