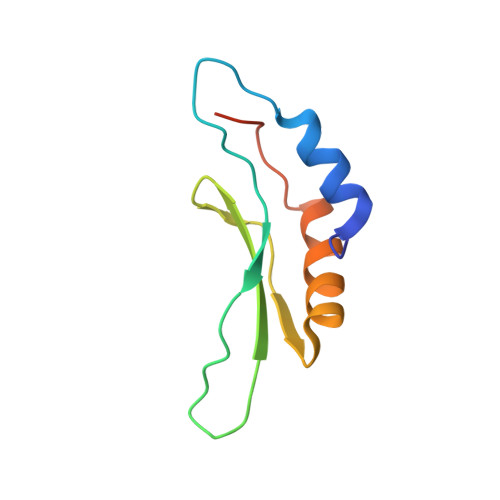
Solution structure of the Drosha double-stranded RNA-binding domain.
Mueller, G.A., Miller, M.T., Derose, E.F., Ghosh, M., London, R.E., Hall, T.M.(2010) Silence 1: 2-2
- PubMed: 20226070
- DOI: https://doi.org/10.1186/1758-907X-1-2
- Primary Citation of Related Structures:
2KHX - PubMed Abstract:
Drosha is a nuclear RNase III enzyme that initiates processing of regulatory microRNA. Together with partner protein DiGeorge syndrome critical region 8 (DGCR8), it forms the Microprocessor complex, which cleaves precursor transcripts called primary microRNA to produce hairpin precursor microRNA. In addition to two RNase III catalytic domains, Drosha contains a C-terminal double-stranded RNA-binding domain (dsRBD). To gain insight into the function of this domain, we determined the nuclear magnetic resonance (NMR) solution structure.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC, USA.