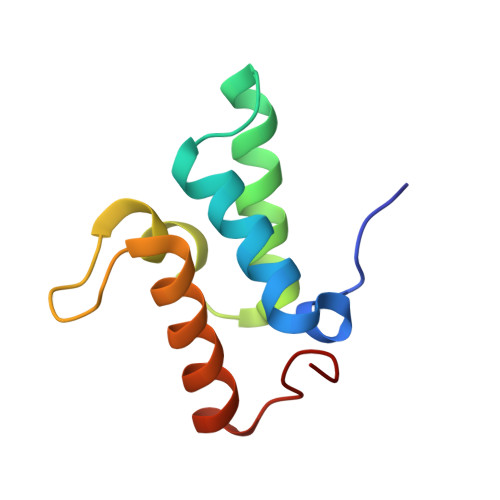
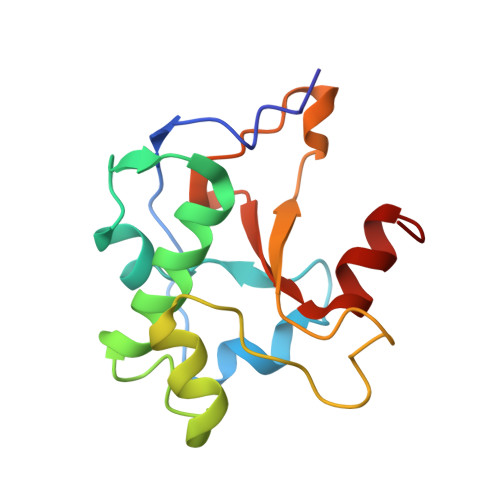
Structure Determination of Protein-Protein Complexes Using NMR Chemical Shifts: Case of an Endonuclease Colicin-Immunity Protein Complex
Montalvao, R.W., Cavalli, A., Salvatella, X., Blundell, T.L., Vendruscolo, M.(2008) J Am Chem Soc 130: 15990-15996
- PubMed: 18980319
- DOI: https://doi.org/10.1021/ja805258z
- Primary Citation of Related Structures:
2K5X - PubMed Abstract:
Nuclear magnetic resonance (NMR) spectroscopy provides a range of powerful techniques for determining the structures and the dynamics of proteins. The high-resolution determination of the structures of protein-protein complexes, however, is still a challenging problem for this approach, since it can normally provide only a limited amount of structural information at protein-protein interfaces. We present here the determination using NMR chemical shifts of the structure (PDB code 2K5X) of the cytotoxic endonuclease domain from bacterial toxin colicin (E9) in complex with its cognate immunity protein (Im9). In order to achieve this result, we introduce the CamDock method, which combines a flexible docking procedure with a refinement that exploits the structural information provided by chemical shifts. The results that we report thus indicate that chemical shifts can be used as structural restraints for the determination of the conformations of protein complexes that are difficult to obtain by more standard NMR approaches.
Organizational Affiliation:
Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK.