Roles of Structure and Structural Dynamics in the Antibody Recognition of the Allergen Proteins: An NMR Study on Blomia tropicalis Major Allergen
Naik, M.T., Chang, C.F., Kuo, I.C., Kung, C.C., Yi, F.C., Chua, K.Y., Huang, T.H.(2008) Structure 16: 125-136
- PubMed: 18184590
- DOI: https://doi.org/10.1016/j.str.2007.10.022
- Primary Citation of Related Structures:
2JMH - PubMed Abstract:
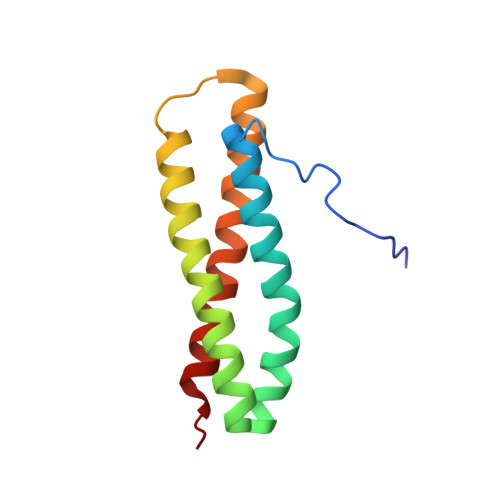
Blo t 5 is the major allergen from Blomia tropicalis mites and shows strong IgE reactivity with up to 90% of asthmatic and rhinitis patients' sera. The NMR solution structure of Blo t 5 comprises three long alpha helices, forming a coiled-coil, triple-helical bundle with a left-handed twist. TROSY-NMR experiments were used to study Blo t 5 interaction with the Fab' of a specific monoclonal antibody, mAb 4A7. The mAb epitope comprises two closely spaced surfaces, I and II, connected by a sharp turn and bearing critical residues Asn46 and Lys47 on one surface, and Lys54 and Arg57 on the other. This discontinuous epitope overlaps with the human IgE epitope(s) of Blo t 5. Epitope surface II is further predicted to undergo conformational exchange in the millisecond to microsecond range. The results presented are critical for the design of a hypoallergenic Blo t 5 for efficacious immunotherapy of allergic diseases.
Organizational Affiliation:
Institute of Biomedical Sciences, Academia Sinica, Taipei 11529, Taiwan.