Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
Igonet, S., Vulliez-Le Normand, B., Faure, G., Riottot, M.M., Kocken, C.H.M., Thomas, A.W., Bentley, G.A.(2007) J Mol Biol 366: 1523
- PubMed: 17229439
- DOI: https://doi.org/10.1016/j.jmb.2006.12.028
- PubMed Abstract:
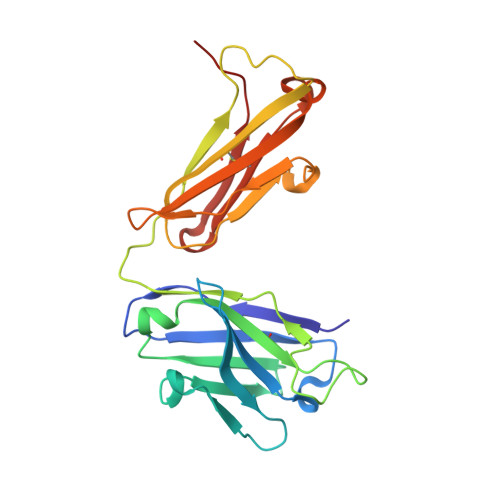
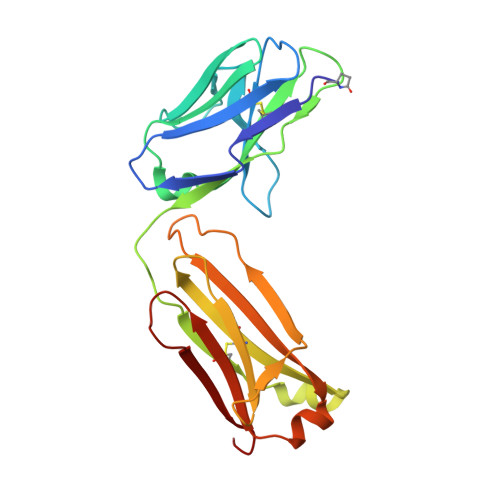
Apical membrane antigen 1 (AMA1) has an important, but as yet uncharacterised, role in host cell invasion by the malaria parasite, Plasmodium. The protein, which is quite conserved between Plasmodium species, comprises an ectoplasmic region, a single transmembrane segment and a small cytoplasmic domain. The ectoplasmic region, which can induce protective immunity in animal models of human malaria, is a leading vaccine candidate that has entered clinical trials. The monoclonal antibody F8.12.19, raised against the recombinant ectoplasmic region of AMA1 from Plasmodium vivax, cross-reacts with homologues from Plasmodium knowlesi, Plasmodium cynomolgi, Plasmodium berghei and Plasmodium falciparum, as shown by immunofluorescence assays on mature schizonts. The binding of F8.12.19 to recombinant AMA1 from both P. vivax and P. falciparum was measured by surface plasmon resonance, revealing an apparent affinity constant that is about 100-fold weaker for the cross-reacting antigen when compared to the cognate antigen. Crystal structure analysis of Fab F8.12.19 complexed to AMA1 from P. vivax and P. falciparum shows that the monoclonal antibody recognises a discontinuous epitope located on domain III of the ectoplasmic region, the major component being a loop containing a cystine knot. The structures provide a basis for understanding the cross-reactivity. Antibody contacts are made mainly to main-chain and invariant side-chain atoms of AMA1; contact antigen residues that differ in sequence are located at the periphery of the antigen-binding site and can be accommodated at the interface between the two components of the complex. The implications for AMA1 vaccine development are discussed.
Organizational Affiliation:
Unité d'Immunologie Structurale, CNRS URA 2185, Département de Biologie Structurale et Chimie, Institut Pasteur, 25 rue du Dr. Roux, 75724 Paris cedex 15, France.