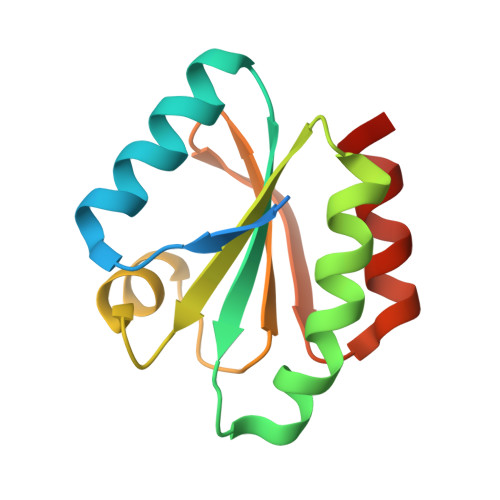
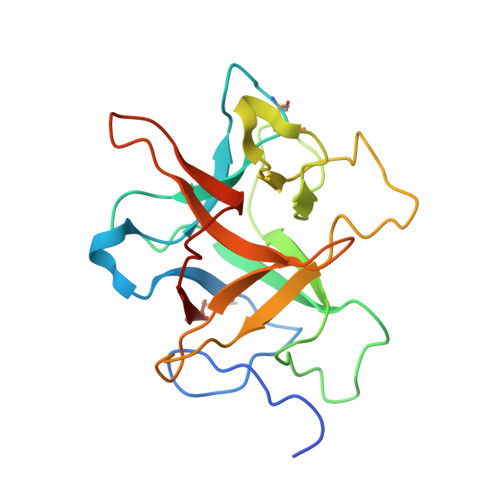
Structural Basis for Target Protein Recognition by the Protein Disulfide Reductase Thioredoxin.
Maeda, K., Hagglund, P., Finnie, C., Svensson, B., Henriksen, A.(2006) Structure 14: 1701
- PubMed: 17098195
- DOI: https://doi.org/10.1016/j.str.2006.09.012
- Primary Citation of Related Structures:
2IWT - PubMed Abstract:
Thioredoxin is ubiquitous and regulates various target proteins through disulfide bond reduction. We report the structure of thioredoxin (HvTrxh2 from barley) in a reaction intermediate complex with a protein substrate, barley alpha-amylase/subtilisin inhibitor (BASI). The crystal structure of this mixed disulfide shows a conserved hydrophobic motif in thioredoxin interacting with a sequence of residues from BASI through van der Waals contacts and backbone-backbone hydrogen bonds. The observed structural complementarity suggests that the recognition of features around protein disulfides plays a major role in the specificity and protein disulfide reductase activity of thioredoxin. This novel insight into the function of thioredoxin constitutes a basis for comprehensive understanding of its biological role. Moreover, comparison with structurally related proteins shows that thioredoxin shares a mechanism with glutaredoxin and glutathione transferase for correctly positioning substrate cysteine residues at the catalytic groups but possesses a unique structural element that allows recognition of protein disulfides.
Organizational Affiliation:
Enzyme and Protein Chemistry, BioCentrum-DTU, Søltofts Plads, Building 224, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark.