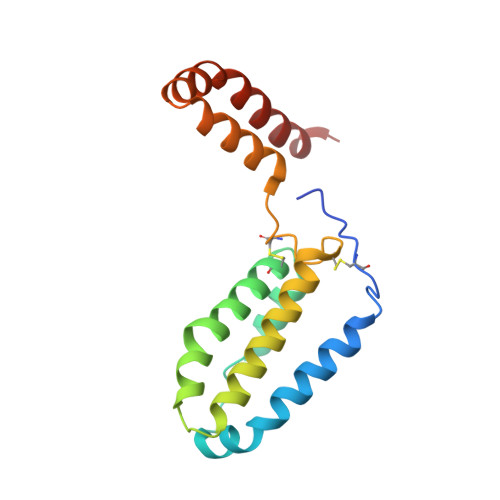
Crystal structure of human interleukin-10 at 1.6 A resolution and a model of a complex with its soluble receptor.
Zdanov, A., Schalk-Hihi, C., Wlodawer, A.(1996) Protein Sci 5: 1955-1962
- PubMed: 8897595
- DOI: https://doi.org/10.1002/pro.5560051001
- Primary Citation of Related Structures:
2ILK - PubMed Abstract:
The crystal structure of human interleukin-10 (IL-10) was refined at 1.6 A resolution against X-ray diffraction data collected at 100 K with the use of synchrotron radiation. Although similar to the IL-10 structure determined previously at room temperature, this low-temperature IL-10 structure contains, in addition, four N-terminal residues, three sulfate anions, and 175 extra water molecules. Whereas the main-chain conformation is preserved, about 30% of the side chains, most of them on the protein surface, assume different conformations. A computer model of a complex of IL-10 with its two soluble receptors was generated based on the topological similarity of IL-10 to interferon-gamma. The contact region between the cytokine and each receptor shows excellent complementarity of polar and hydrophobic interactions, suggesting that the model is generally correct and should be useful in guiding mutagenesis experiments.
Organizational Affiliation:
Macromolecular Structure Laboratory, NCI-Frederick Cancer Research and Development Center, ABL-Basic Research Program, Frederick, Maryland 21702, USA.